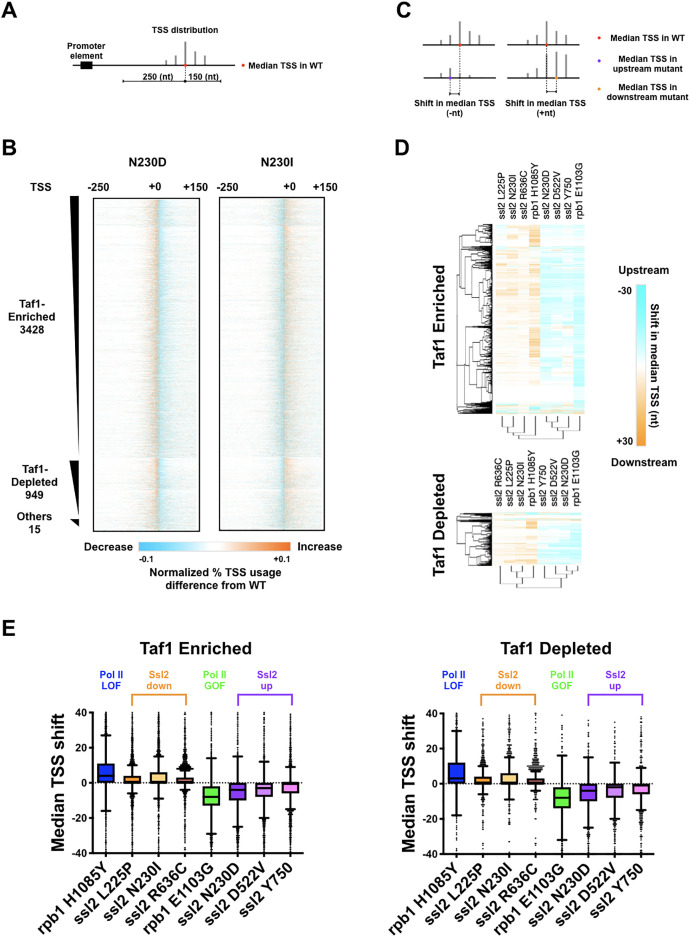
Figure 4. ssl2 alleles shift transcription start site (TSS) distribution genome-wide.
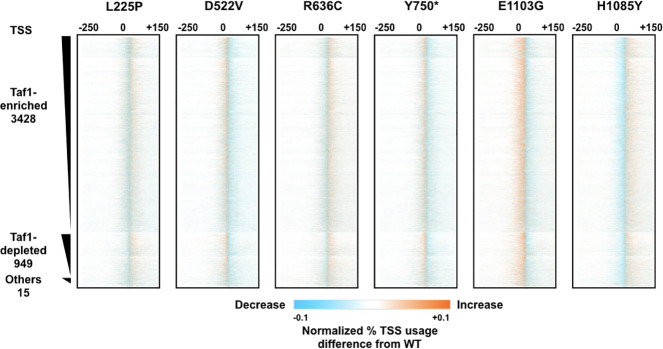
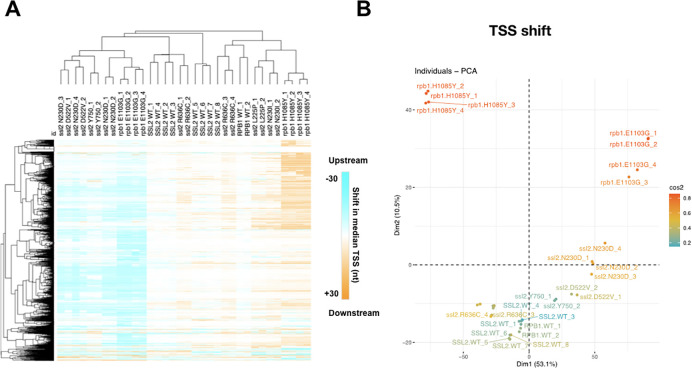
(A) Schematic indicating TSS distribution within a promoter window defined by median of the wild-type (WT) TSS distribution. (B) Heatmaps of TSS normalized read differences between WT and ssl2 mutants within defined promoter windows. Promoter classes defined by enrichment or depletion of Taf1 were rank-ordered according to total reads in WT. The promoter-normalized read differences between mutant and WT are shown by a color scheme ranging from cyan (negative)-orange (positive). (C) Schematic illustrating median TSS upstream or downstream shift in a mutant relative to WT. (D) Heatmap and hierarchical clustering of median TSS shifts for ssl2 or rpb1 mutants for Taf1-enriched or -depleted promoter classes (promoters as in (B)). The shift of median TSS is indicated by cyan (upstream) or orange (downstream). (E) TSS shifts in ssl2 mutants are less strong than in Pol II mutants. Median TSS shifts across promoters, regardless of promoter class are statistically distinguished from zero at p < 0.0001 for all mutants (Wilcoxon signed rank test).