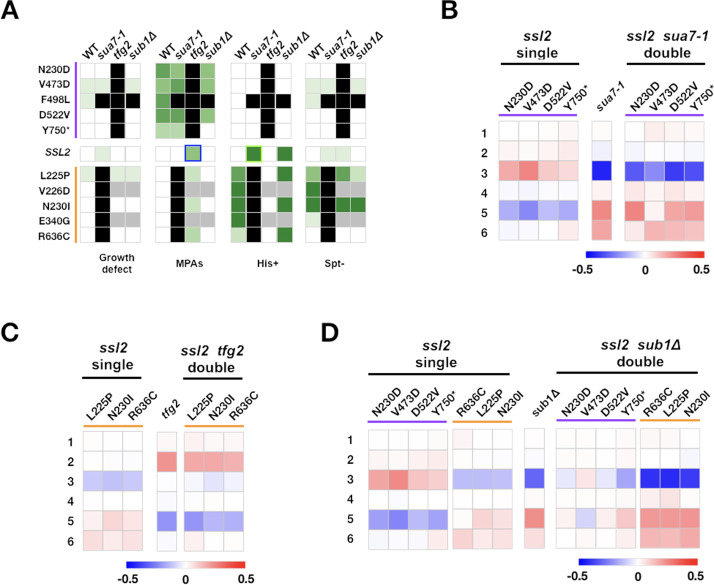
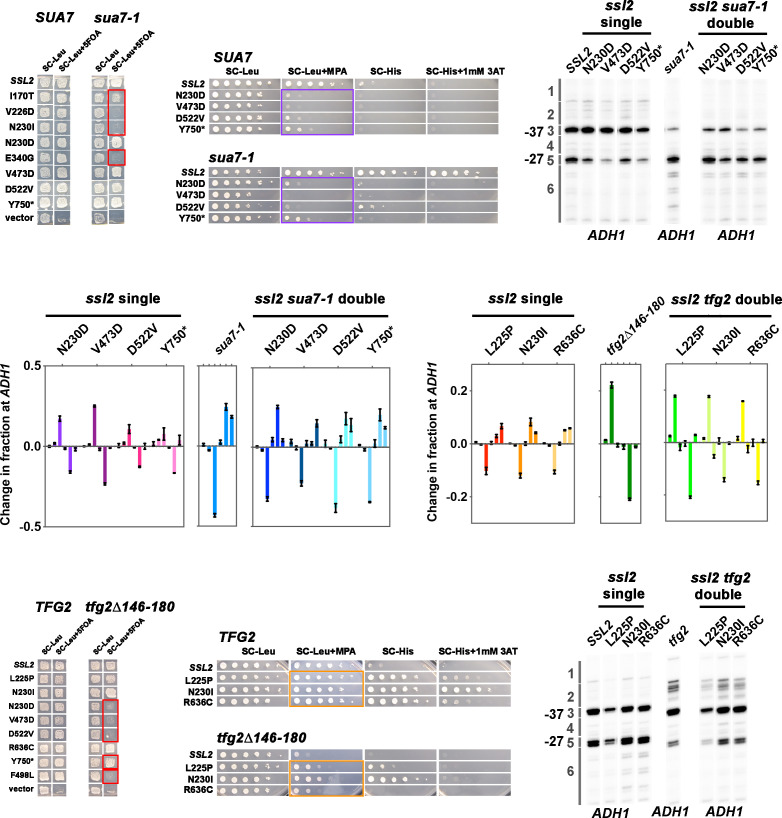
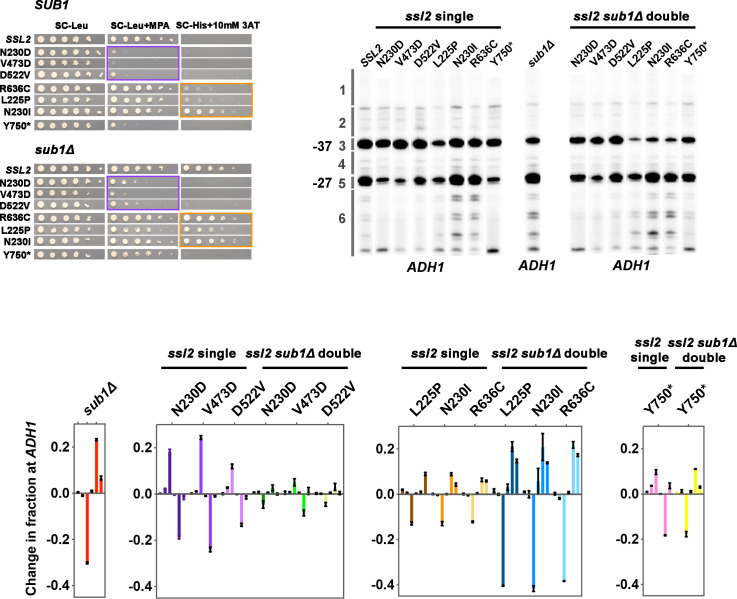
(A) Lethality is broadly observed between ssl2 downstream shifting alleles and the sua7-1 downstream shifting allele. Patch assay showing that six downstream shifting alleles I170T, V226D, N230I, E340G, L225P, and R636C show synthetic lethality phenotypes when combined with sua7-1 allele (ssl2 I170T and sua7-1 double mutant is very sick on 5FOA plate but cannot be propagated to single colonies). Four ssl2 upstream shifting alleles N230D, V473D, D522V, Y750* show normal growth phenotypes when sua7-1 is incorporated. One ssl2 upstream shifting allele F498L showed unique (lethality) genetic interactions compared to other ssl2 upstream shifting alleles (normal growth). This allele additionally shows lethality with both TFIIF and sub1∆ alleles as discussed in the corresponding sections. (B) ssl2 upstream shifting alleles show epistasis with the sua7-1 downstream shifting allele for MPAS phenotypes. Spot assay shows that four upstream shifting ssl2 alleles’ strong MPAS phenotypes (N230D, V473D, D522V, and Y750*) are almost completely unaffected when sua7-1 allele, which shows a His+ phenotype by itself, is incorporated. Primer extensions shown are representative of ≥3 independent biological replicates. (C) Double mutants of ssl2 upstream shifting alleles and sua7-1 downstream shifting allele shift TSS distribution downstream. Primer extension results show that ssl2 alleles’ effect on shifting TSS distribution upstream is almost completely reversed to downstream shifting after incorporation of sua7-1 allele, with only slight additive effect seen in double mutants’ TSS usage. Primer extensions shown are representative of ≥3 independent biological replicates. (D) Quantification of primer extension detected TSS usage at ADH1 in ssl2, sua7-1 single or double mutants. Bars are averages ± standard deviation of ≥3 independent biological replicates. (E) Lethality is observed between ssl2 upstream shifting alleles and the tfg2∆146–180 upstream shifting allele. Patch assay illustrates that upstream shifting alleles N230D, V473D, D522V, and F498L show synthetic lethality phenotypes when combined with tfg2∆146–180 allele (ssl2 Y750* and tfg2∆146–180 double mutant shows moderate level of growth on 5FOA plate, however, shows strong sickness when propagated to single colonies). Three ssl2 downstream shifting alleles L225P, N230I, and R636C show normal growth phenotypes when tfg2∆146–180 is incorporated. (R636C in double mutant shows slight sicker growth phenotype than as single mutant.) (F) ssl2 downstream shifting alleles show both epistasis and additive effects with tfg2∆146–180 downstream shifting allele. Spot assay shows that the double mutants of ssl2 downstream alleles and tfg2∆146–180 allele show MPAS phenotypes; however, double mutants’ MPAS phenotypes are not as strong as observed in single tfg2∆146–180 allele. (G) Double mutants of ssl2 downstream shifting alleles and tfg2∆146–180 upstream shifting allele shift TSS distribution upstream. Primer extension results show that ssl2 alleles’ effect on shifting TSS distribution upstream is almost completely reversed to upstream shifting after incorporation of tfg2∆146–180 allele. Primer extensions shown are representative of ≥3 independent biological replicates. (H) Quantification of primer extension detected TSS usage at ADH1 in ssl2, tfg2∆146–180 single or double mutants. Bars are averages ± standard deviation of ≥3 independent biological replicates.