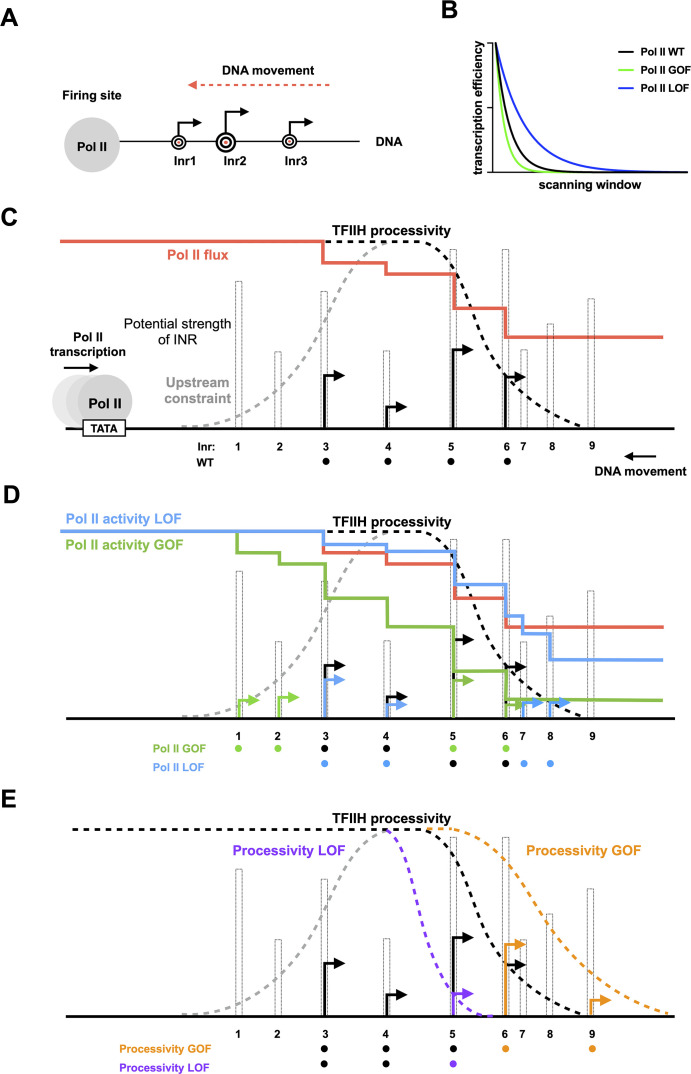
Figure 9. Model for interaction between initiation efficiency and scanning processivity.
(A) The ‘Shooting Gallery’ model. The polymerase II (Pol II) active site controls initiation efficiency, that is, ‘the rate of firing’. TFIIH controls the rate and extent of scanning, that is, ‘the speed of target passage and number of targets reached’. (B) Reduction in relative transcription start site (TSS) usage as scanning Pol II initiates. As Pol II (wild-type [WT]) scans from upstream to downstream, successful initiation at upstream positions will reduce the amount of Pol II continuing to scan downstream. Increasing initiation efficiency at each position as is predicted for increased Pol II catalytic activity will result in a more rapid decrease in observed initiation from upstream to downstream. Conversely, reducing initiation efficiency at each position will flatten observed TSS distribution because more Pol II will reach downstream positions. (C) TSS distributions during promoter scanning in the ‘Shooting Gallery’ model. The TSS distribution (black arrows) of a promoter window can be affected by Pol II catalytic activity, preinitiation complex (PIC) scanning rate and processivity, TSS strength, Pol II flux, and additional observed (upstream limitation on initiation too close to PIC assembly) or potential (downstream limitation through chromatin structure) constraints. (D) Effects of Pol II catalytic activity on TSS distributions. Increased Pol II catalytic activity increases the efficiency of upstream TSSs that are encountered by Pol II and decreases the usage of downstream TSSs due to quickly reduced Pol II flux (changes indicated as green arrows). Decreased Pol II catalytic activity decreases TSS efficiency of upstream TSSs encountered by Pol II and increases apparent TSS usage at downstream sites due to failed upstream initiation, resulting in a downstream shifted TSS distribution within a window determined by PIC scanning potential (changes shown as blue arrows). (E) Effects of altered scanning processivity on TSS distributions. Increased processivity alleles are hypothesized to increase the probability of Pol II scanning further downstream if Pol II flux remains, thus expanding the scanning window and allowing Pol II usage of downstream TSSs if Pol II flux is not limiting (orange TSS). In contrast, decreased processivity will limit Pol II scanning downstream, truncating the distribution of observed TSSs (purple TSS).