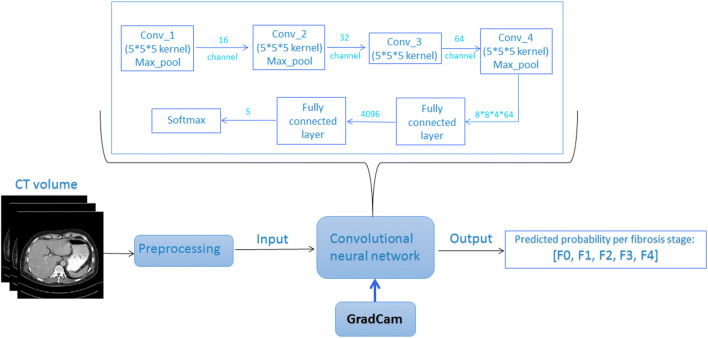
Fig. 2.
Overall scheme of liver fibrosis staging by deep learning. The computed tomography (CT) scan was first pre-processed by tissue windowing and standardized to [0,1]. Then, 32 consecutive slices of the 3D segmented liver were randomly selected as a patch per training iteration to feed into the convolutional neural network. The network put out the array of predicted probabilities at fibrosis stage (F0–F4). During testing of the trained liver fibrosis staging network, Grad-Cam was integrated between the third convolutional layer and the final convolutional layer to generate the maps showing the location of the network’s focus. Abbreviations: CT: computed tomography; Conv: convolutional layer; Max_pool: maximum pooling layer; GradCam: Gradient-weighted Class Activation Mapping; 5*5 kernel: a kernel with size [5] is used to extract features in the convolutional layer; channel: number of kernels applied in between convolutional layers; Softmax: softmax activation function