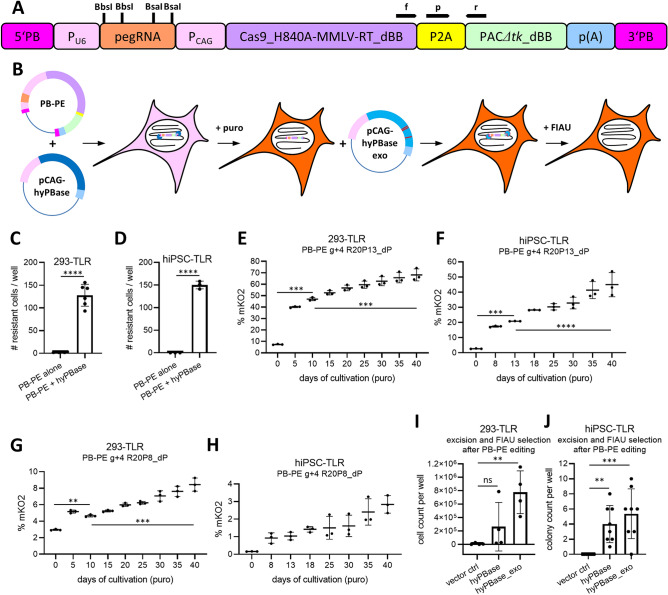
Figure 4.
Proof-of-principle for a piggyBac prime editing (PB-PE) system allowing for enrichment of gene edited cells in human bulk cell populations. (A) Unscaled schematic of PB-PE vector. Cas9_H840A-MMLV-RT_dBB and PACΔtk_dBB are modified versions of PE22 and puroΔtk13, respectively, where all Bbsl and Bsal sites were deleted. Approximate binding sites of PB-PE-specific qRT-PCR primers (f, r) and probe (p) are indicated as black tapered lines. (B) Graphic overview of a PB-PE based gene editing approach. (C, D) Calculated colony formation efficiency after puromycin selection of 293-TLR (C) and hiPSC-TLR (D) cells transfected with PB-PE transposon with and without hyPBase transposase. (E) FACS analysis for mKO2 gene corrected 293-TLR cells which were PEI-transfected with PB-PE g+4 R20P13_dP transposon and pCAG-hyPBase at different days of puromycin selection. (F) FACS analysis for mKO2 gene corrected hiPSC-TLR cells edited with PB-PE g+4 R20P13_dP transposon at different days of puromycin selection. (G, H) FACS analysis for mKO2 gene corrected 293-TLR (G) and hiPSC-TLR (H) cells edited with PB-PE g+4 R20P8_dP transposon at different days of puromycin selection. (I) Cell count per well of PB-PE g+4 R20P13_dP-edited 293-TLR cells, after transposon excision by hyPBase or hyPBase_exo (excision-optimized) transposase and FIAU counter-selection. (J) Colony count per well of PB-PE g+4 R20P13_dP-edited hiPSC-TLR cells, after transposon excision by hyPBase or hyPBase_exo transposase and FIAU counter-selection. Vector ctrl: cells transfected with L.CGIP (see Supplementary Methods). Data are represented as ± SD from n = 3 (D–H), n = 4 (I), n = 6 (C) or n = 8 (J) biological replicates and significance was calculated using 1-way ANOVA with Tukey's post-test except for (C) and (D) where unpaired t-test was used (****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01; ns: p > 0.05).