Abstract
Objectives:
Neutrophil extracellular traps (NETs) are extracellular lattices composed of nucleic material bound to neutrophil granule proteins. NETs may play pathogenic roles in development and severity of autoimmune diseases such as systemic lupus erythematosus (SLE), at least in part, through induction of type I interferon (IFN) responses via externalization of oxidized immunostimulatory DNA. A distinct subset of SLE proinflammatory neutrophils (low density granulocytes; LDGs) displays enhanced ability to form proinflammatory NETs that damage the vasculature. We assessed whether NET-bound RNA can contribute to inflammatory responses in endothelial cells and the pathways that mediate this effect.
Methods:
Expression of newly-synthesized and total RNA was quantified in HC and lupus NETs. The ability of endothelial cells to take up NET-bound RNA and downstream induction of type I IFN responses was quantified. RNAs present in NETs were sequenced and specific small RNAs were tested for induction of endothelial type I IFN pathways.
Results:
NETs extruded RNA that was internalized by endothelial cells and this was enhanced when NET-bound nucleic acids were oxidized, particularly in lupus LDG NETs. Internalization of NET-bound RNA by endothelial cells was dependent on endosomal TLRs and the actin cytoskeleton and induced type I IFN stimulated genes (ISGs). This ISG induction was dependent on NET-associated miR-let7b, a small RNA expressed at higher levels in LDG NETs, which acted as a TLR7 agonist.
Conclusion:
These results highlight underappreciated roles for small RNAs externalized in NETs in the induction of proinflammatory responses in vascular cells, with implications to lupus vasculopathy.
Introduction.
Systemic lupus erythematosus (SLE) is an autoimmune syndrome characterized by development of autoantibodies that target nucleic acids or proteins binding to nucleic acids, pleiotropic clinical manifestations, immune complex (IC) deposition in various organs and acute and chronic inflammation (1, 2). Endogenous nucleic acids released during various forms of cell death have been considered important sources of autoantigens in SLE, leading to chronic stimulation of innate and adaptive immune responses. Recognition of self-nucleic acids by endosomal TLRs on various immune cells is considered an important step in the pathogenesis of SLE, promoting autoantibody and IC formation and the production of type I interferons (IFNs), with pathogenic effects (3).
In addition to apoptosis and necrosis as putative sources of externalized nuclear autoantigens, there is another form of cell death whereby extracellular traps extruded by neutrophils contain modified DNA. Dysregulation on formation and clearance of these neutrophil extracellular traps (NETs) may play important roles in generating and exposing modified autoantigens to the immune system, and in amplifying inflammatory responses in SLE and other autoimmune conditions (4). SLE is characterized by presence of a distinct subset of proinflammatory neutrophils, low-density granulocytes (LDGs), endowed with enhanced capacity to form NETs (5, 6) in a mitochondrial reactive oxygen species-dependent manner (7, 8). Oxidation of genomic and mitochondrial DNA during NET formation amplifies immunogenicity of this nucleic acid and promotes type I IFN responses through engagement of the cyclic GMP-AMP synthase/stimulator of IFN genes (cGAS-STING) pathway (8, 9). NETs also damage the endothelium and may play pathogenic roles in the development of vasculopathy and premature atherosclerosis characteristic of SLE (4, 10).
While mitochondrial DNA and chromatin are present in NETs, it is unclear if additional nuclear components with potential immunomodulatory capabilities become externalized and modified during this process, and the role that they may play in immune dysregulation and tissue injury. RNA, particularly small RNAs, are increasingly described for their central roles in promoting cross-talk between cells and their immunomodulatory and proinflammatory activities (11). RNAs can be delivered to other cells inside microvesicles or exosomes and, following uptake, can regulate gene expression in target cells (12). Recently, RNAs present in NETs have been associated with perpetuation of inflammation and subsequent propagation of NET formation in psoriasis skin lesions (13), while NET-bound microRNA miR-142 induces TNF expression by macrophages in vitro (14).
We hypothesized that NETs serve as carriers of RNAs that may regulate endothelial cell (EC) responses and that differential RNA expression in lupus NETs may induce exacerbated proinflammatory responses in the endothelium. To test this hypothesis, we assessed whether RNA present in NETs could be internalized by ECs. We also characterized differential expression of miRNAs in healthy control (HC) and SLE NETs and their effects on EC biology and type I IFN responses.
Material and methods:
Cell lines.
Human aortic ECs (HAECs; catalog#: CC-2535) and human dermal microvascular ECs (HMVECs; catalog#: 2543) were purchased from Lonza, seeded over gelatin (Sigma Aldrich, #G1890)-coated cell culture containers in MCDB 131 (Gibco, ThermoFisher Scientific, #10372019) or EBM-2 Endothelial Cell Growth Basal Medium (Lonza, #CC-3156) containing EGM-2 Endothelial SingleQuots kit (Lonza, #CC4176). Cells were used after no more than 10 passages.
Isolation of neutrophils and NETs.
Low- and normal-density neutrophils were isolated from peripheral venous blood of SLE subjects and HCs, as described (8, 15). SLE subjects met revised ACR criteria for this disease (16) and were enrolled in protocol NIH#94-AR-0066. Healthy volunteers were recruited at the Blood Bank Clinical Center, NIH. All subjects signed informed consent. Neutrophils (2 × 106 cells/ml RPMI) were allowed to form spontaneous NETs for 1.5 h at 37°C, followed by addition of 10 units/ml RNase free-DNase (ThermoFisher Scientific, #00672727). Some of the NET preparations were induced in the presence of 5 μl 5-ethynyl uridine (EU) reagent from the Click-It RNA Alexa Fluor 488 Imaging kit (ThermoFisher Scientific, #C10329), to label newly-synthesized RNA. NETs were collected after 30 min, further separated from cells by centrifugation (5 min at 5,000 rpm), followed by filtering through 0.45 um syringe filters to exclude whole cells (GE Healthcare Life Sciences, #6901–2504). NET aliquots were stored at −80°C until use.
Immunofluorescence microscopy.
Neutrophils were seeded onto chambered coverslips (Lab-Tek ThermoFisher Scientific, #155383) at a density of 100,000 cells/ml, allowed to attach for 1.5 h, followed by fixation in 4% paraformaldehyde overnight at 4°C. Cells were washed with PBS, permeabilized with Triton X100 0.2 % for 10 min, washed and incubated with Click-It reagents, following manufacturer’s instructions. Cells were blocked in gelatin 0.5% for 15 min, then incubated with various fluorochrome-conjugated antibodies after labeling with Click-It. All anti-human antibodies were from abcam and included anti-LL-37 (1:250, mouse monoclonal #ab80895), anti-neutrophil elastase (1:500, rabbit polyclonal #ab21595), anti-histone H2A (1:250, rabbit polyclonal #ab18255), and anti-high mobility group box 1 (HMGB1) (1:250, rabbit polyclonal #10829–1-AP). Secondary antibodies, used at 1:250 dilution, were from ThermoFisher Scientific and included: donkey anti mouse Alexa 488 (#A-21202) and donkey anti rabbit Alexa 555 and 488 (#A-32773 and A-21206). Hoechst 33342 (ThermoFisher Scientific; # H3570) was used at 1:1,000 dilution to stain nuclear and extracellular DNA fibers, in conjunction with the secondary antibody. For neutrophils and NETs, incubations were done overnight at 4°C for primary antibody and for 2 h at room temperature with the secondary antibody and Hoechst. ProLong Gold antifade reagent (ThermoFisher Scientific, #P36930) was used to mount the samples that were then imaged in a Zeiss LSM780 confocal microscope. Total protein measurement in NETs was done with BCA kit (ThermoFisher Scientific; #23225) and 50 ug/ml were used per experiment. For analysis of microvesicles in NETs using Image J, images were examined in Photoshop and we applied the setting Photoshop>filter>stylize>find edges to estimate the mean size of vesicles containing newly-synthesize RNA in NETs
Quantification of NET-bound newly-synthesized and total RNA.
Newly-synthesized RNA in neutrophils and NETs was labeled using the Click-It RNA imaging kit after incubating cells for 1.5 h with 5-ethynyl uridine (EU; 5 μl/ml), following manufacturer’s instructions. Additional experiments labeled NET-bound RNA using SYTO RNASelect green dye (ThermoFisher Scientific, #S32703) or Exo-Red exosome Glow (SBI System Biosciences, #EXOR100A-1). SYTO RNASelect green dye was added to NETs for the duration of the incubations (1 μl/ml). Exo-Red exosome Glow dye was added to 100 μl/ml of NETs for 20 min at 37°C, followed by addition of 200 μl TCA for 40 min on ice. Suspensions were centrifuged for 10 min at 14,000 rpm and the pellet containing labeled NETs was resuspended in 500 μl PBS and stored at −80°C. Staining of total RNA in EC lines was performed by incubating cells with SYTO RNA Select green dye (1 μl/ml media) for 1 h at 37°C.
In vitro oxidation of NETs and determination of oxidized nucleotide (8-OHG) content.
Spontaneously formed NETs from healthy volunteer neutrophils were oxidized in vitro following an adapted protocol. (17). In brief, one ml NETs was mixed with 25 uM cytochrome c (Sigma Aldrich, #C2867) and 1 mM hydrogen peroxide in HEPES pH 7.3 for 1 h at 37°C. The reaction was stopped with 5mM EDTA and oxidized NETs were stored at −80°C. Negative controls were NETs without addition of cytochrome c and hydrogen peroxide. Oxidation of nucleic acids was confirmed by an 8-OHG immunoblot and also performed in LDGs without in vitro oxidation. In brief, 5 μl NET suspensions were attached to a nitrocellulose membrane strip and allowed to dry. After blocking by incubation in 10% BSA and successive washes in 0.05% PBS-Tween 20, the nitrocellulose membrane was incubated overnight at 4°C with anti-8-OHG mouse monoclonal antibody (1:500, abcam; #ab62623), washed with 0.05% PBS-Tween 20 and incubated with donkey anti mouse IRDye 800 CW LI-COR (1:10,000 #926–32212). The membrane was scanned and imaged in an Odyssey CLx imager.
Assessment of NET-bound RNA uptake by ECs.
EC lines were seeded onto gelatin chambered coverslips (density of 25,000 cells/ml). Semiconfluent cells were incubated with EU-containing NETs (50 ug/ml protein) for 3 h, washed in PBS and fixed in 4% paraformaldehyde overnight at 4°C. To assess the uptake of NET-derived newly-synthesized RNA containing EU into ECs, cells were treated using the Click-It newly-synthesized RNA labeling procedure, permeabilized for 10 min with Triton X100 0.2%, washed and blocked in gelatin 0.5% for 30 min. Immunostaining used the protocol described above for neutrophils, to quantify internalization of neutrophil newly-synthesized RNA. In additional experiments aimed at quantifying total RNA uptake by ECs, NETs were labeled with Exo-Red exosome Glow and then incubated with ECs in 96-well plates in the presence or absence of RNase1 (20 U/ml), RNaseA (0.1 mg/ml) and RNaseH (50 U/ml) (ThermoFisher Scientific, EN0601, EN0531 and EN0201, respectively); TLR7/9 inhibitor and respective control at 4 uM final concentration (Invivogen, ODN TTAGGC(A151) and ODN TTAGGG control); chloroquine 20 uM final concentration (Sigma Aldrich, # C6628) or cytochalasin D 2 ug/ml final concentration (Sigma Aldrich, #C8273). Experiments were performed in triplicate. Cells were washed in PBS and nuclei were stained with Hoechst to quantify cell numbers. Fluorescence (extinction/emission: 460/650 and 361/497 nm, respectively) was measured in a Synergy HTX BioTek plate reader.
Isolation and sequencing of NET-bound small RNAs and use of small RNA inhibitors and mimics.
NET-bound RNA was isolated using ExoMir kit (BIOO Scientific, #5145). For each sample, 1 μg eluted RNA was subjected to limited alkaline hydrolysis in a 15 μl buffer of 10 mM Na2CO3 and 10 mM NaHCO3 (pH 10.3) at 60°C for 10 min. The partially hydrolyzed RNA was dephosphorylated with 10 U calf intestinal phosphatase (New England Biolabs) in a 50 μL reaction of 100 mM NaCl, 50 mM Tris-HCl (pH 7.9) at 25°C, and 10 mM MgCl2, 1 mM DTT, 3 mM Na2CO3, and 3 mM NaHCO3, at 37°C for 1 hr. The resulting RNA was re-phosphorylated with 10 U T4 polynucleotide kinase (NEB) in a 20 μl reaction of 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT, and 1 mM ATP at 37°C for 1 hr. Fragments of 19–35 nt were converted into barcoded small RNA cDNA libraries, as previously described (18), and sequenced on an Illumina HiSeq 2500 instrument. Adapters were trimmed using cutadapt (http://journal.embnet.org/index.php/embnetjournal/article/view/200/458). Alignment and annotation were performed as described (19). Sequencing reads were mapped to human genome hg19 with Bowtie/0.12.8. Small RNA expression values (RPKM) were calculated using Partek/6.6, with genomic annotations generated using HOMER (20). ANOVA was performed with Partek/6.6. The sequence data files are deposited in GEO, accession number GSE160143.
The miRCURY LNA miRNA power inhibitor and negative control against miR-let7b was from Qiagen (YI04102235 and YI00199006-DCA) and used at 50 nM final concentration. The miR-let7b MISSION microRNA mimic was from Sigma Aldrich (HMI0007) and was also used at 50 nM final concentration.
Immunoprecipitation of HGMB1-bound miR-let7b and quantification by qPCR.
Immunoprecipitation of NET-bound HMGB1 was performed by incubating 1 ml control or SLE NETs with 1 ug anti-HMGB1 (rabbit-antihuman #10829–1-AP) overnight at 4°C. The suspension was incubated with protein A conjugated agarose beads (GE Healthcare, # 17127901; 50 μl diluted at 2x in PBS) for 2 h at room temperature. After 3 washes in water, agarose pellets were stored at −80°C. Elution of RNA from immunoprecipitated HMGB1 was done as described (21). After elution, the RNA was precipitated, and each sample resuspended in 15 μl water. The miR-let-7b qPCR was performed using the hs_let-7b_1 miScript Primer Assay Qiagen (#MS00003122) with Highspec buffer, 12 μl of template for the cDNA synthesis and following the kit instructions for qPCR.
Gene expression analysis by qPCR.
Gene expression in HAECs was quantified using specific taqman primers probes (Thermofisher Scientific, GAPDH: Hs02786624_g1, IFNA5: Hs04186137_sH, IFI44: Hs00951349_m1, IRF7: Hs01014809_g1) after isolation of the RNA using the Direct-zol RNA miniprep kit (Zymo Research: #R2050). The qPCR was done following a protocol described by us (22).
Results.
NETs express newly-synthesized RNA that is internalized by endothelial cells.
The first indication that RNA was present in NETs from lupus neutrophils was obtained using Nanodrop and confirmed by Bioanalyzer quantification (Table S1). Results showed that the amount of RNA in SLE versus HC-derived cells was not significantly different. RNA present in NETs was predominantly small size RNA; a representative Bioanalyzer electrophoretogram report for RNA isolated from lupus normal dense granulocytes (NDGs) NETs is shown in Figure 1A. To confirm that RNA was present in NETs in a more refined manner we used the Click-It technology (23) to label and track de novo synthesized RNA. Newly-synthesized RNA in neutrophils was effectively labeled following a 2 h incubation in the presence of modified ribonucleotide 5-ethynyl uridine (EU), which is incorporated into newly-synthesized RNA and can be subsequently fluorescently labeled. Labeled newly-synthesized RNA was detected in vesicle-like structures in HC NDGs (in green, Figure S1A) and SLE LDGs (Figure S1B) before active NET formation. By confocal microscopy, we observed that newly-synthesized RNAs were present in well-defined vesicle-like structures within NETs (Figure 1B–C). The mean size of these vesicles was 1.99 + 0.16 um in diameter. Overall, these results indicated that NETs externalize newly synthesized RNA.
Figure 1: RNA is detected in NETs, and newly-synthesized RNA in NETs colocalizes in vesicle-like structures.
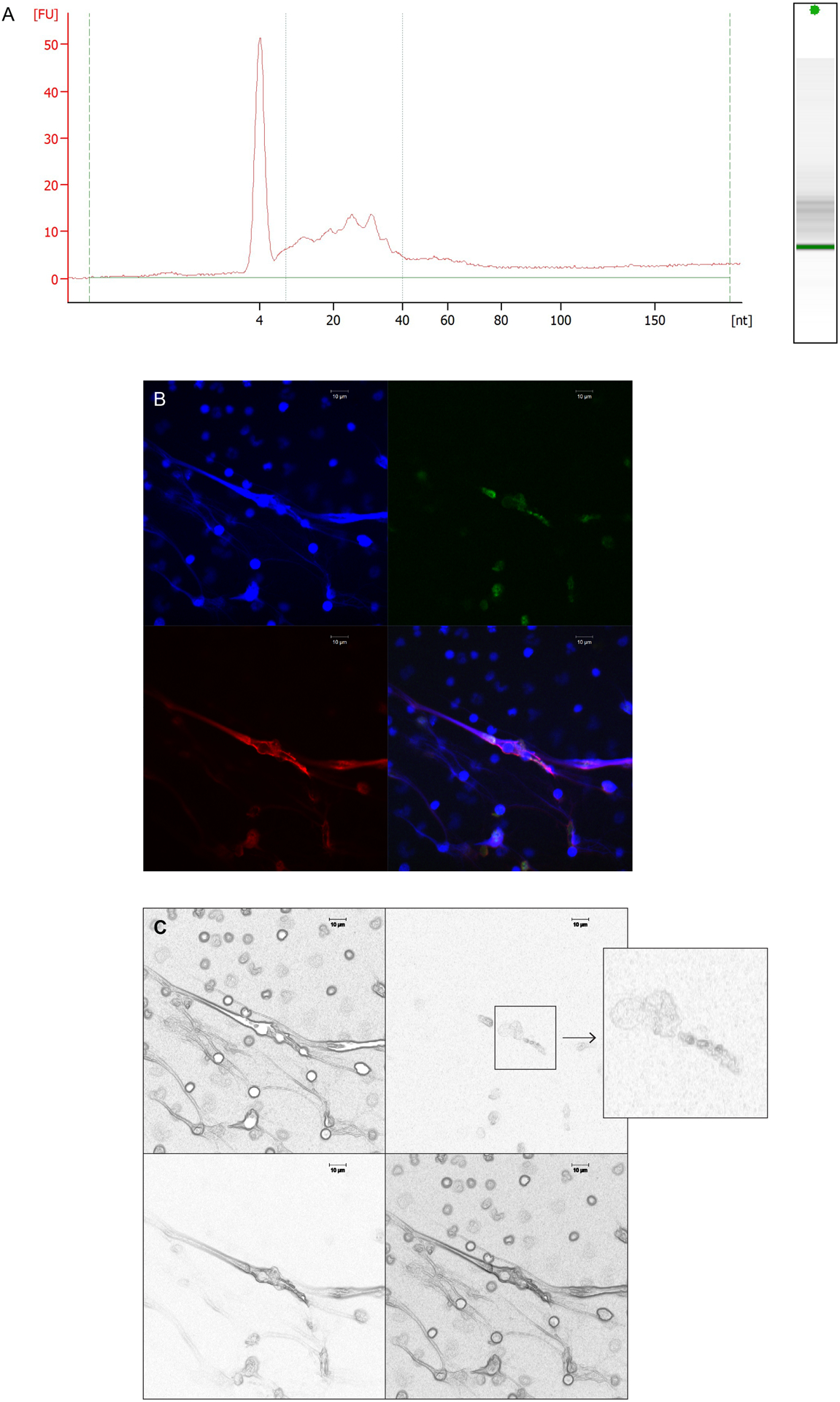
A) Small RNAs are the predominant RNA species quantified in NETs, as shown in a Bioanalyzer electrophoretogram report of RNA from a representative SLE NDG NET sample. B) Confocal microscopy image of SLE LDGs undergoing NET formation. The newly-synthesized RNA (green) in NETs is contained in vesicle-like structures that colocalize with extracellular DNA (blue) and neutrophil elastase (red). Magnification is 63x and the scale bar shown corresponds to 10 um. C) Image in B was processed using Photoshop>filter>stylize>find edge, then the image was transformed to black and white. The insert is a magnification of the area where the newly-synthesized RNA shows a vesicle like morphology.
We tracked the uptake of NET-associated newly-synthesized RNA by ECs. HAECs were incubated for 2.5 h with lupus-derived NETs carrying EU-labeled RNA, permeabilized, and stained to detect internalized newly-synthesized RNA. As shown in Figure 2, newly-synthesized RNA derived from NETs was internalized by HAECs, detected in their cytoplasm in vesicle-like structures (Figures 2A–C) and colocalized with neutrophil granule proteins neutrophil elastase (Figure 2A) and neutrophil derived LL-37 (Figure 2B); both molecules are externalized during NET formation and were quantified to track colocalization of RNAs with granule proteins contained within NETs. NET-derived newly-synthesized RNA also trafficked into HAEC nuclei, as displayed in the fluorescence profile graphs (Figure 2A). Coincubation of NETs with RNase A for the duration of the assay attenuated uptake of newly-synthesized NET-bound RNA by HAECs (Figure 2C). Furthermore, the DNA/protein NET scaffold appeared to be required for efficient uptake of NET-bound RNA because naked newly-synthesized RNA purified from NETs was not readily taken up by HAECs (Figure S2A); however, adding 1 uM recombinant LL37 (as used by (24)) together with the RNA, further enhanced RNA internalization (Figure S2B). Overall, these results indicated that newly-synthesized RNA present in NETs can be internalized by ECs. Furthermore, after Image J image analysis of several images from different experiments, the amount of internalized newly-synthesized RNA from NETs is higher when using NETs derived from lupus patients compared to HCs (Figure S3).
Figure 2: Newly-synthesized RNA from NETs gets internalized by HAECs.
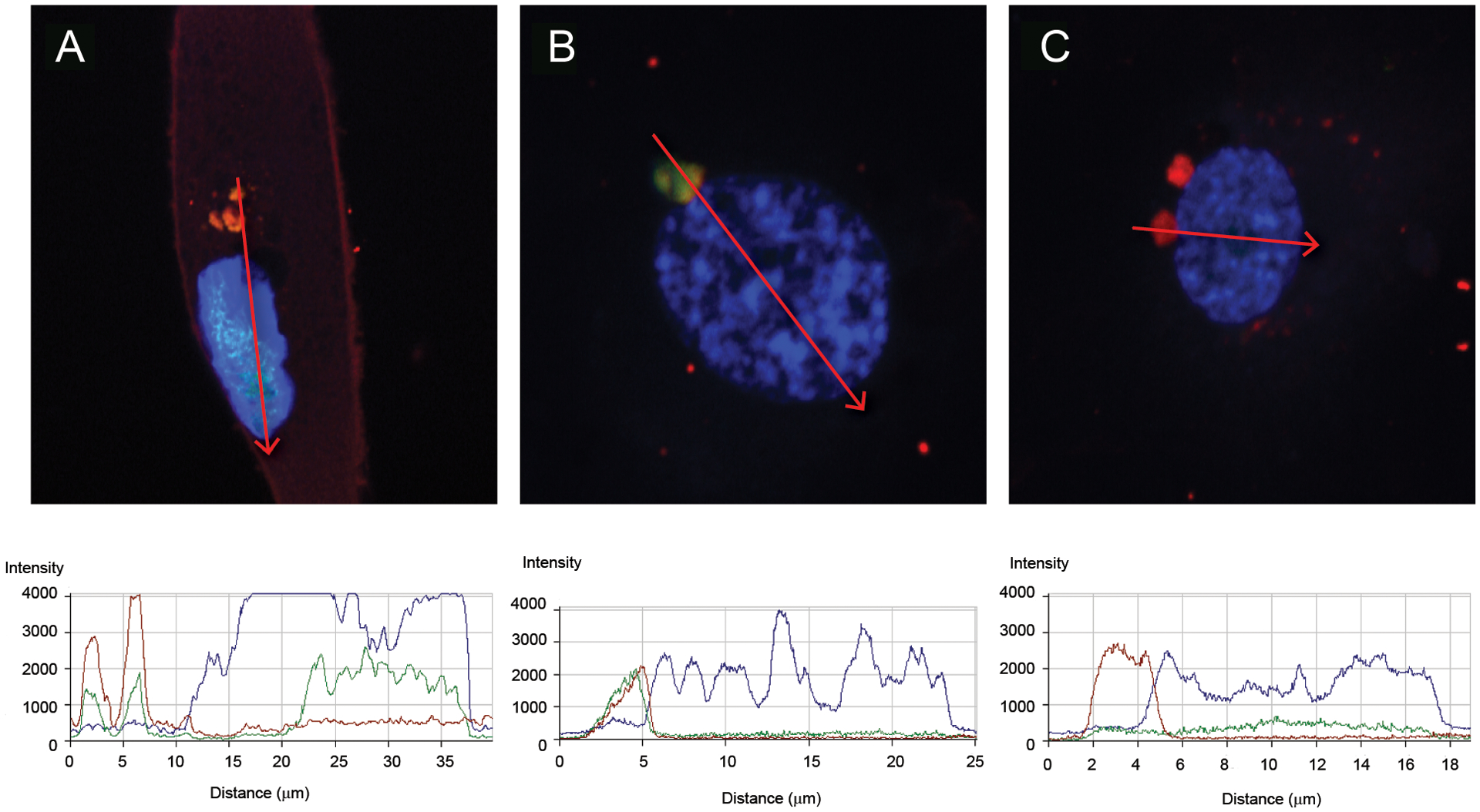
Analysis of the uptake of newly-synthesized RNA from NETs into HAECs by laser confocal microscopy. Shown are representative images of permeabilized HAECs with intracellular newly-synthesized RNA labeled in green, DNA in blue (Hoechst), and neutrophil elastase (A and C) or LL-37 (B) in red. In C, experiment was performed in the presence of RNase A. For each profile image, the fluorescence for each fluorochrome was quantified in the region of the red arrow displayed, and the respective fluorescence quantification is shown in the bottom graphs. The images are representative of at least three different experiments using different SLE and HC samples. Magnification are 126x.
The oxidative status of NETs modulates RNA uptake by ECs.
Previous studies suggested that the level of DNA oxidation in NETs modulates the induction of type I IFN responses in target cells and that lupus LDG NETs contain higher amounts of oxidized nucleic acids than NETs generated by other types of stimulation (8). To explore if oxidation status of nucleic acids present in NETs modulates the ability of target cells to internalize NET-bound newly-synthesized RNA, we oxidized in vitro spontaneously generated NETs purified from HC neutrophils that were labeled with EU to identify newly-synthesized RNA. Following an in vitro oxidation procedure using cytochrome c (17), we confirmed by dot blot the presence of oxidized nucleic acids using an antibody that recognizes 8-hydroxyguanosine. Both spontaneously formed SLE-derived LDG NETs and in vitro oxidized spontaneously generated HC NETs displayed enhanced amounts of oxidized nucleic acids compared to non-oxidized HC NETs (Figure 3A). Degree of uptake of newly-synthesized RNA by HAECs correlated with level of nucleic acid oxidation in NETs (Figure 3B–C) and was the highest for lupus LDG-derived NETs that carried the most elevated levels of nucleic acid oxidation (Figures 3D). To confirm these data, nuclei-associated green fluorescence representative of internalized newly-synthesized NET-associated RNA was quantified using image J (Figure S3A). To exclude that green fluorescence detected in HAECs was due to incorporation of traces of free EU carried by NETs, HAECs were incubated with free EU. The faint diffuse fluorescence pattern detected with free EU was different from the intense fluorescence pattern from newly-synthesized RNA labeled in NETs (Figure S3B). To confirm findings, we used another dye that stains total RNA, SYTO RNA Select green dye, which also labels HAEC nucleoli (Figure S3C). Internalized NET-bound newly-synthesized RNA displayed enhanced localization in nucleoli. Overall, these observations indicate that ability of NET-bound RNA to be internalized by ECs is dependent on the oxidation status of the nucleic acids in NETs, with RNA bound to lupus NETs being more readily internalized.
Figure 3: NET derived-newly-synthesized RNA uptake by endothelial cells increases with degree of nucleic acid oxidation in NETs.
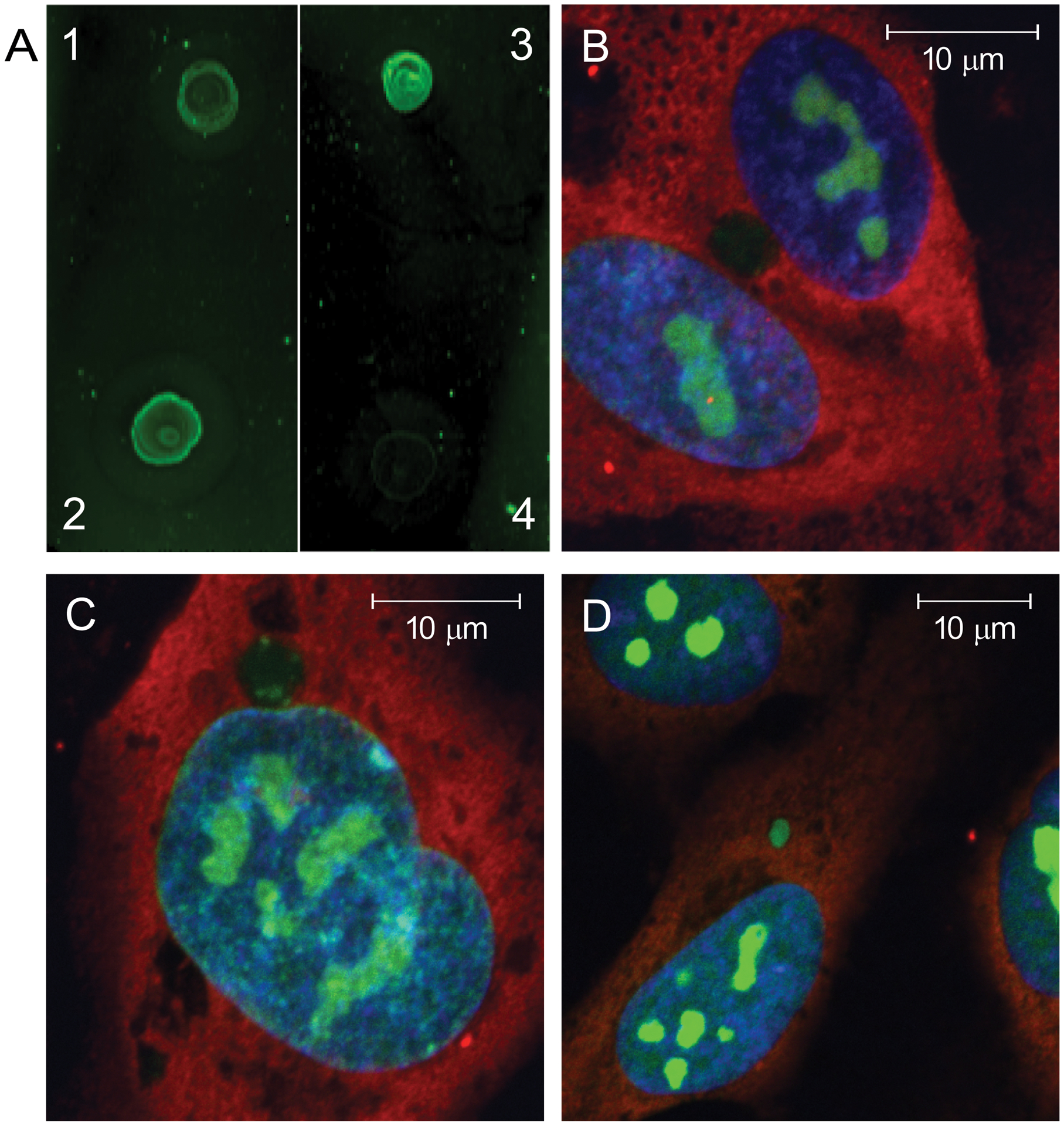
NETs spontaneously derived from healthy volunteer NDGs were oxidized in vitro using cytochrome c. To determine the level of nucleic acid oxidation, a dot blot using anti-8-hydroxyguanosine antibody is displayed in A, depicting HC NETs before (1) and after (2) in vitro oxidation. Controls include NETs from a lupus LDG subject without added in vitro oxidation (3), and vehicle alone without NETs (4). HAECs were incubated with non-in vitro oxidized control NETs (B), in vitro oxidized control NETs (C), or non-in vitro oxidized lupus LDG NETs (D) for 3 hours and newly-synthesized RNA uptake was imaged. The confocal images display internalized newly-synthesized RNA (green), DNA (blue) and neutrophil elastase (red). The images are representative of three independent experiments. Nuclei-associated green and blue fluorescence from images B, C, D, and images from additional experiments were quantified using Image J (Figure S2). Magnifications are: B and C 126x; D 97x. Scales bars are 10 um in each image.
Uptake of NET-bound total RNA by ECs depends on the actin cytoskeleton and endosomal TLRs.
To characterize the mechanisms by which NET-bound RNA is internalized by ECs, we labeled total RNA in NETs with Exo-Red Glow. This dye is based on the chemistry of acridine orange designed for staining nucleic acids contained in exosomes, interacts with RNA and DNA but emits fluorescence in distinct wavelengths (red for RNA and green for DNA). The total RNA-labeled NETs were incubated with HAECs in a plate assay to quantify by fluorometry both total RNA uptake and relative number of cells by staining nuclei with Hoechst. The ability of ECs to internalize NET-bound total RNA was significantly reduced by preincubation of these cells with cytochalasin D (an inhibitor of actin cytoskeleton polymerization) and by chloroquine (an inhibitor of endosomal acidification/endosomal TLRs) (Figure 4A) (25). Uptake of RNA was also inhibited by a specific TLR7/9 inhibitor, suggesting a role for endosomal TLRs in internalization of NET-bound RNA and consistent with previous observations that NET internalization by other cells, such as synovial fibroblasts, is dependent on the function of these TLRs (26). To further assess the type of NET-bound RNA that may be preferentially internalized by ECs, we tested different RNAses (RNAse H, RNase 1 and RNase A). Both RNase 1 and RNase A act similarly on single stranded RNAs, particularly under high salt conditions (27). In contrast, RNase H preferentially targets RNA-DNA hybrids. Addition of either RNases or HAEC-derived non-labeled RNA to digest or compete with NET-bound RNAs, respectively, significantly decreased the uptake of NET-bound labeled RNA by HAECs (Figure 4A). Different RNases had variable effects in reducing ability of EC to internalize NET-bound RNA, suggesting that NETs contain diverse types of RNAs with potential to be internalized by HAECs. The most effective inhibitor was RNase H, followed by RNase 1 and RNase A. Representative laser confocal images are shown in Figure 4B. Overall, these results indicate that ECs internalize RNA through mechanisms involving the actin cytoskeleton and endosomal TLRs.
Figure 4: Uptake of NET-bound total RNA by endothelial cells depends on the actin cytoskeleton and endosomal TLRs.
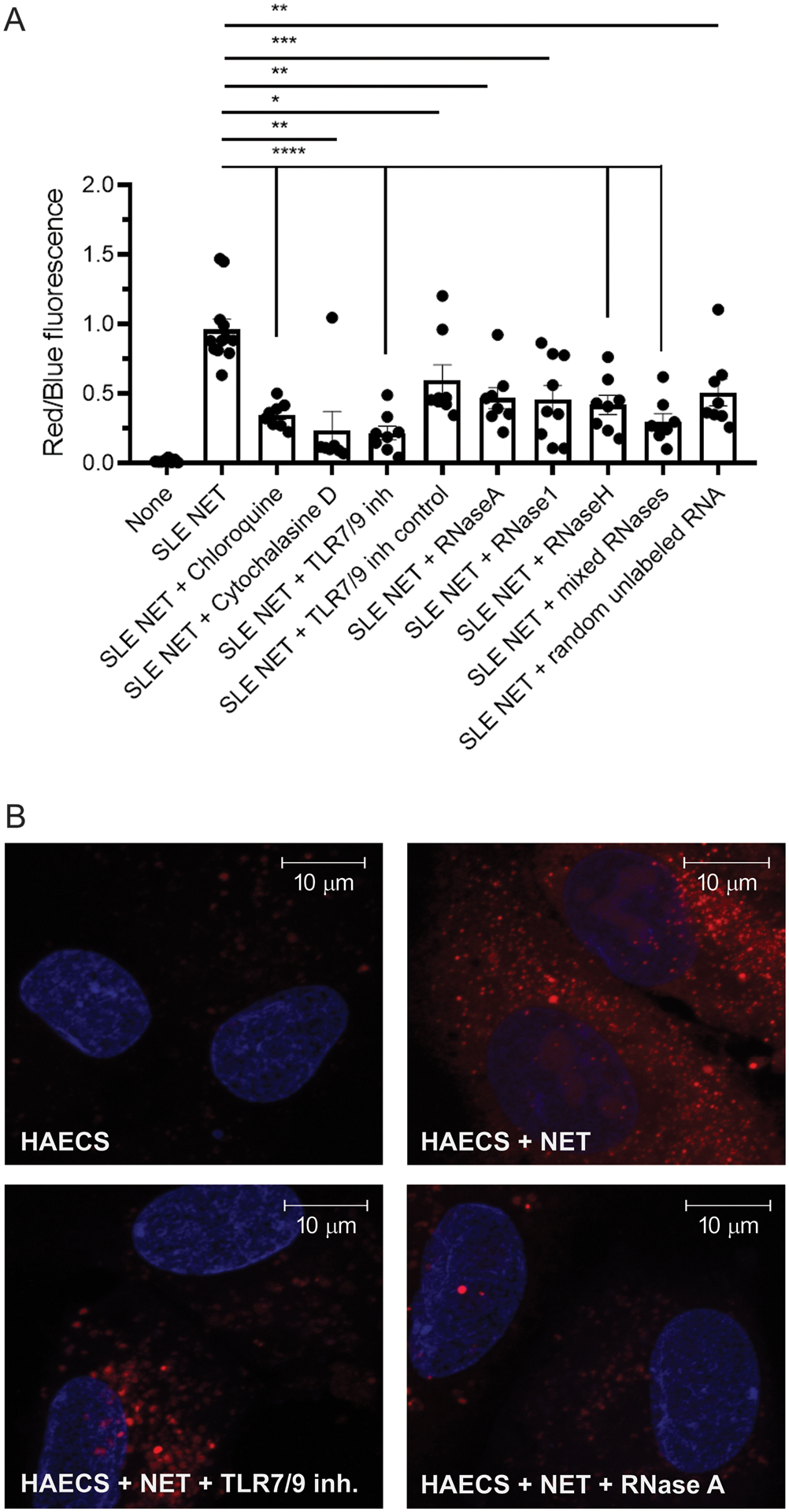
Hoechst (blue) was used to quantify number of cells in A and to label nuclei in B. Shown in (A) is the ratio between Exo Glow red fluorescence staining internalized total RNA derived from NETs and blue fluorescence from Hoechst staining DNA both quantified by plate assay and fluorometry. (B) Representative images of similarly stained HAECs with total RNA in NETs stained with the Exo Glow red dye by laser confocal. Upper images: HAECs alone (left); HAECs plus NETs (middle); HAECs plus NETs in the presence of TLR7/9 inhibitor (right). Lower images: HAECS plus NETs and RNase A (left); HAECs plus NETs and RNase 1 (middle); HAECs plus NETs and all RNases combined (right). Bar graphs represent the mean ± SEM of three independent experiments. Statistical significance was analyzed by Mann-Whitney U test *: p<0.05; ** p<0.01. Magnifications in images are 126x and scale bars in images correspond to 10 um.
Characterization of NET-bound RNAs.
Identifying that some NET-associated RNAs are contained in vesicle like structures (Figure 1) led us to characterize methods to purify the NET-bound RNA derived from HC and SLE neutrophils, by using a kit designed for RNA isolation from exosomes. To exclude remaining whole cells from the NET analyses, suspensions were filtered through a 0.45 um pore size filter before RNA isolation procedure, as indicated in materials and methods. We then sequenced NET-bound small RNAs (Table S2). Hierarchical clustering and principal component analyses (PCA) (Figure 5A–B) revealed significant differences in small RNA expression in NETs from SLE NDGs and LDGs (yellow and red) and HC NDGs (blue). Among the small RNAs sequenced, most of them were more abundant in spontaneous NETs from HC NDGs (Table 2S and Figure 5C), compared to SLE derived NETs. NETs from SLE NDGs were enriched in miR-7704 and miR-24–1, compared to NETs from HC NDG and SLE LDGs (Figure 5C). The most abundant small RNAs in LDG NETs were miR-let7b and miR-27b (Figure 5C), when compared to SLE and HC NDG-derived NETs. Therefore, subsequent work focused on their putative role in modulating EC biology.
Figure 5: Small RNAs present in NETs characterize by sequencing.
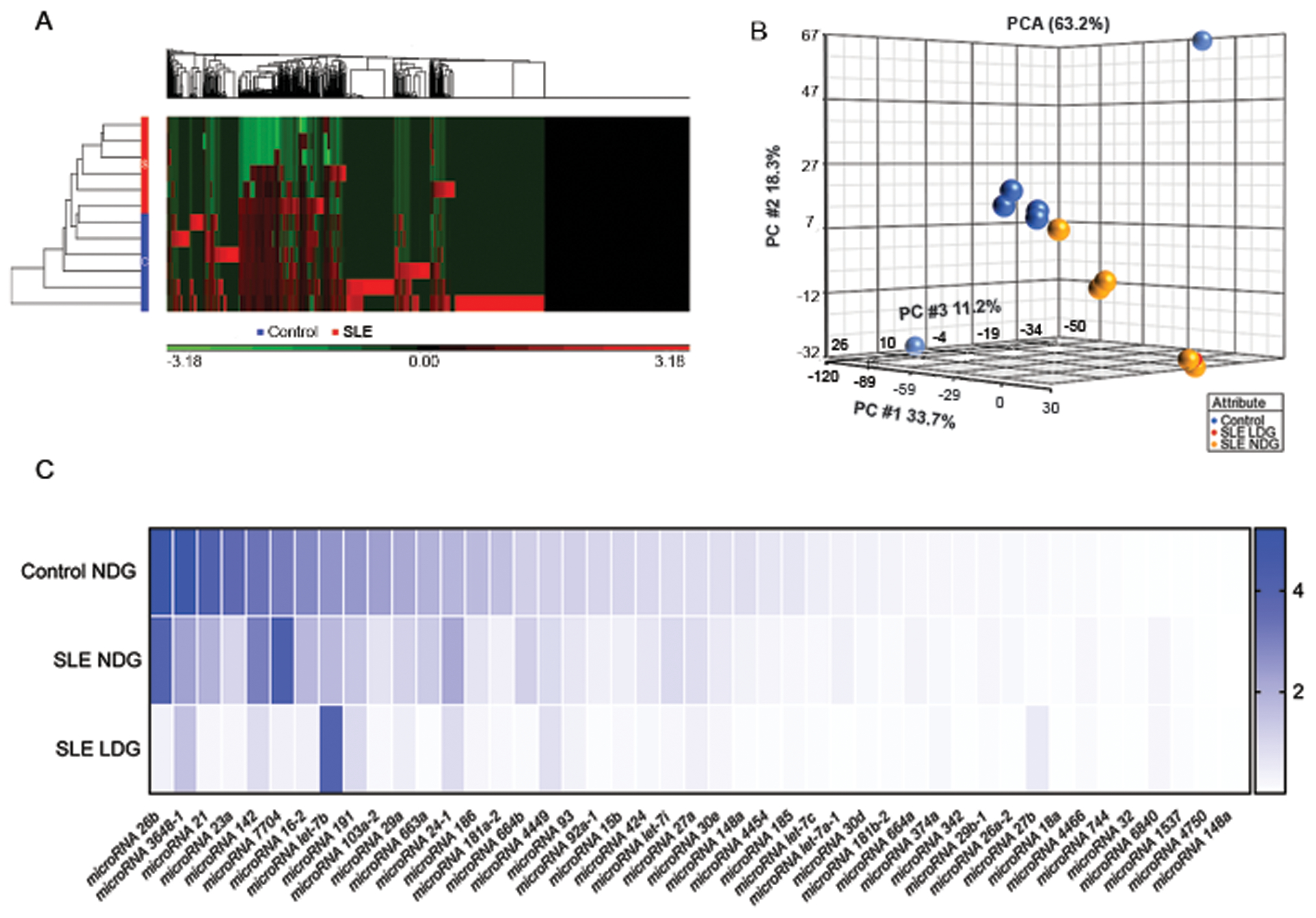
A) Hierarchical clustering after Partek analysis of the small RNA sequencing. B) Principal component analysis of small RNA sequences comparing NETs from HC NDGs (blue), SLE LDGs (red), and SLE NDGs (yellow). C) Heat map graph displays the expression of selected small RNAs from NETs purified from HC NDGs, SLE NDGs, or SLE LDGs. The gene expression in log2(rpkm) was normalized to the housekeeping small RNA gene RNU6ATAC. The small RNA sequencing analysis in NETs was done using 12 different samples (6 from lupus patients and 6 from HCs).
NET-bound small RNA miR-let7b promotes type I IFN responses in ECs.
We hypothesized that small RNAs present in NETs may regulate proinflammatory responses in ECs. NETs can induce type I IFN responses in myeloid cells, at least in part, through oxidation of nucleic acids that trigger the cGAS-STING pathway (8). To test if NETs promote type I IFN responses in ECs, which induces proinflammatory responses in vasculature, we incubated HAECs with spontaneously generated NETs from SLE NDGs and LDGs and from HC NDGs for 6 (Figure 6A) or 24 hours (Figure 6B–D), and quantified type I IFN mRNAs and type I IFN stimulated genes (ISGs). Only NETs derived from SLE NDGs and LDGs but not NETs derived from HC neutrophils, significantly enhanced expression of IFNA, IFNB and the ISGs IFI44 and IRF7 in ECs (Figures 6A–D).
Figure 6: miR-let7b present in NETs induces type I IFN responses in endothelial cells.
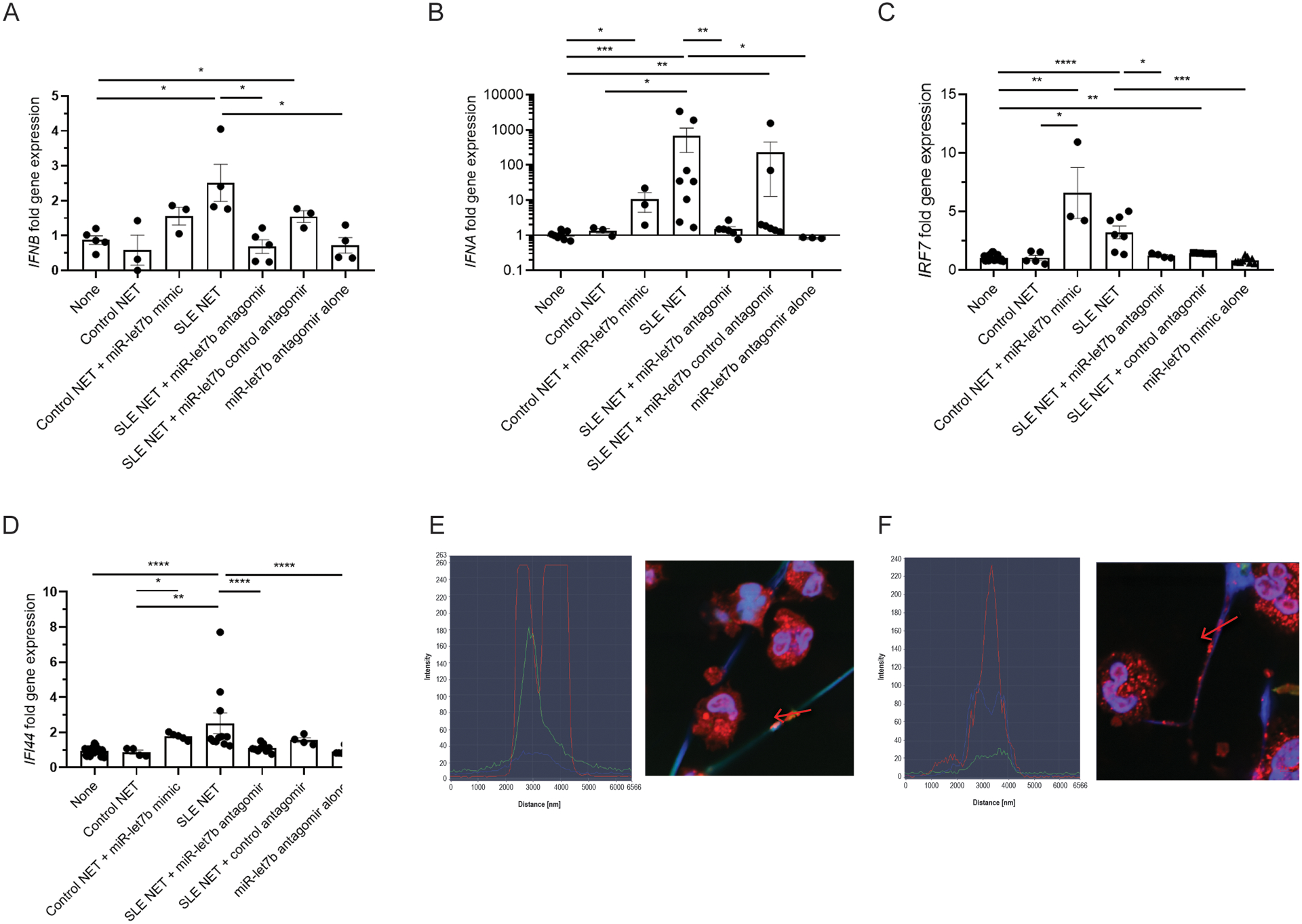
A) IFNB, B) IFNA, C) IFI44, and D) IRF7 gene expression in HAECs quantified after 6 h (A) and 24 h (B, C, D) coincubation with NETs from SLE (average LDG and NDG) or HC NDGs (control NET) in the presence or absence of the antagomir for miR-let7b and control antagomir. Results also display the effect of an miR-let7b mimic added to HC NETs or the antagomir alone. In NETs, HMGB1 colocalizes with total RNA (E and F). NETs and nuclei are stained with Hoechst (blue), HMGB1 (red) and total RNA (SYTO dye green). Representative images are from SLE (E) LDG or (F) NDG cells and NETs. Left portion of the images depicts plots of profile fluorescence quantification through the red arrow depicted on the NET (included in each image). Results are representative of 3 independent experiments.
To assess if type I IFN responses induced in ECs by lupus NETs are driven, at least in part, by specific small RNAs, we evaluated putative candidates. As mentioned above, miR-let7b was the highest expressed small RNA in LDG NETs. It was previously implicated in induction of ISGs in rheumatoid arthritis synovial macrophages and in nociceptor neurons, as it has a GU rich sequence that directly activates TLR7 (28, 29). We incubated HAECs with NETs in the presence or absence of a specific antagomir (antisense oligonucleotide inhibitor) against miR-let7b or a control antagomir (oligonucleotide sequence with <70% homology to any sequence in any organism in NCBI and miRBase databases), and measured IFNB, IFNA, and ISGs. As shown in Figures 6A–D, the miR-let7b antagomir (but not the control antagomir) significantly inhibited expression of these IFN genes, suggesting that miR-let7b contributes to induction of type I IFN responses in ECs triggered by lupus NETs. To further confirm this effect, we incubated HAECs with or without HC NDG-derived NETs in the presence or absence of exogenously added miR-let7b mimic and then quantified gene expression. The miR-let7b mimic added to HC NETs significantly induced IFN gene expression in HAECs (Figures 6A–D).
A putative candidate by which miR-let7b interacts with TLR7 is HMGB1, a histone-like alarmin described in NETs (30) that forms complexes with miR-let7b, leading to cell toxicity in TLR-7 dependent models (31). Indeed, total RNA colocalized with HMGB1 in NETs and this was enhanced in lupus LDG-derived NETs when compared with NDG-derived NETs (Figures 6E–F). Following immunoprecipitation with an antibody against HMGB1, we quantified remnant HMGB1-associated miR-let7b by qPCR. Indeed, HMGB1-associated miR-let7b was more abundant in SLE LDG NETs than autologous NDG NETs (Figure S4). These data suggest that small RNA miR-let7b present in NETs promotes type I IFN responses in ECs and that this phenomenon is enhanced in lupus-derived NETs.
Discussion
We report that NETs contain small RNAs that, upon uptake and internalization by ECs, induce type I IFN responses. This process is enhanced by NET-associated oxidized nucleotides that are significantly increased in SLE LDG NETs. This resultant enhanced oxidation of nucleotides in lupus NETs correlated with the ability of ECs to internalize NET-bound RNA. Previous reports indicated that oxidation of cell membranes increases the ability of cells to internalize extracellular cargoes, by reducing lipid hydrophobicity (32). Oxidation of NETs, either in vivo through neutrophil-derived ROS or in vitro as shown in this work, may modify vesicle-associated lipids that contain RNAs and enhance their uptake by ECs.
The alarmins LL-37 and HMGB1 have RNA binding activity and are present in NETs (6, 13, 33). We confirmed that ECs exposed to NETs take up LL-37 that colocalizes with neutrophil-derived newly-synthesized RNA. This is consistent with previous studies that have reported that exogenous LL-37 can enter target cells and disrupt nuclear membranes during NET formation (34). Furthermore, both HMGB1 and LL-37 have been associated with ability of pDCs to take up exogenous DNA from NETs (30, 35, 36) and enhance type I IFN synthesis. Also, previous evidence indicates that extracellular LL-37 can bind to extracellular self-RNA and transfer it to DCs’ endosomes to activate type I IFN responses through TLR7/TLR8. As such, the presence of both RNA and LL-37 in NETs may lead to the creation of stable complexes that protect RNA from degradation and promote immunomodulatory and vasculopathic activity that is enhanced in the context of SLE NETs (37, 38).
Among the small RNAs contained in the NETs, miR-let7b was described to display interferogenic activity on pDCs (37), given its intrinsic ability to act as a natural ligand of TLR7 (28, 31). Recently, miR-let7b was characterized as an endogenous TLR7 agonist involved in inflammation propagation in psoriatic arthritis (39). Induction of type I IFN responses in ECs has been linked to development of vasculopathy, and atherothrombosis, particularly in SLE, through pleiotropic effects on vascular repair, inflammation and coagulation (40–43). As such, the induction of endothelial interferogenic responses induced by small RNAs bound to NETs may be highly conducive to vascular damage. The roles of endogenous miR-let7b in endothelial function are complex, and may also involve protecting these cells from oxidation (44–47). Therefore, future studies should comprehensively address how this NET-bound small RNA modulates overall vascular health in SLE and other inflammatory diseases. Of note, other components of NETs can induce endothelial damage, including histones (48–50) and matrix metalloproteinases (10). Therefore, dissecting the precise role that these different NET components play in overall development of vasculopathy should be characterized in future studies, to define the best therapeutic target.
Overall, we report that NETs are a source of intracellular RNAs that impact EC biology. These effects are particularly driven by small RNAs and their interactions with NET-derived alarmins. These observations are relevant not only for vascular damage induced in SLE but, also, for other conditions associated with NET formation and vascular damage such as sepsis, atherothrombosis and cancer. Future studies should assess if targeting specific RNAs present in NETs could have therapeutic implications in these potentially devastating conditions.
Supplementary Material
Acknowledgement:
We are grateful to Ms. Daniela Aguero for expert support in preparation of figures.
Funding:
Supported by the Intramural Research Program at NIAMS/NIH (ZIA AR041199).
Footnotes
There are no conflicts of interest
References:
- 1.Tsokos GC. Systemic Lupus Erythematosus. New England Journal of Medicine. 2011;365(22):2110–21. [DOI] [PubMed] [Google Scholar]
- 2.Rahman A, Isenberg DA. Systemic Lupus Erythematosus. New England Journal of Medicine. 2008;358(9):929–39. [DOI] [PubMed] [Google Scholar]
- 3.Barrat FJ, Crow MK, Ivashkiv LB. Interferon target-gene expression and epigenomic signatures in health and disease. Nature Immunology. 2019;20(12):1574–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan MJ. Neutrophils in the pathogenesis and manifestations of SLE. Nat Rev Rheumatol. 2011;7(12):691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny MF, Yalavarthi S, Zhao W, Thacker SG, Anderson M, Sandy AR, et al. A Distinct Subset of Proinflammatory Neutrophils Isolated from Patients with Systemic Lupus Erythematosus Induces Vascular Damage and Synthesizes Type I IFNs. The Journal of Immunology. 2010;184(6):3284–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villanueva E, Yalavarthi S, Berthier CC, Hodgin JB, Khandpur R, Lin AM, et al. Netting Neutrophils Induce Endothelial Damage, Infiltrate Tissues, and Expose Immunostimulatory Molecules in Systemic Lupus Erythematosus. The Journal of Immunology. 2011;187(1):538–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmona-Rivera C, Kaplan M. Low-density granulocytes: a distinct class of neutrophils in systemic autoimmunity. Semin Immunopathol. 2013;35(4):455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lood C, Blanco LP, Purmalek MM, Carmona-Rivera C, De Ravin SS, Smith CK, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nature medicine. 2016;22(2):146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehrke N, Mertens C, Zillinger T, Wenzel J, Bald T, Zahn S, et al. Oxidative Damage of DNA Confers Resistance to Cytosolic Nuclease TREX1 Degradation and Potentiates STING-Dependent Immune Sensing. Immunity. 2013;39(3):482–95. [DOI] [PubMed] [Google Scholar]
- 10.Carmona-Rivera C, Zhao W, Yalavarthi S, Kaplan MJ. Neutrophil extracellular traps induce endothelial dysfunction in systemic lupus erythematosus through the activation of matrix metalloproteinase-2. Annals of the rheumatic diseases. 2015;74(7):1417–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herster F, Bittner Z, Archer NK, Dickhöfer S, Eisel D, Eigenbrod T, et al. Neutrophil extracellular trap-associated RNA and LL37 enable self-amplifying inflammation in psoriasis. Nature Communications. 2020;11(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linhares-Lacerda L, Temerozo JR, Ribeiro-Alves M, Azevedo EP, Mojoli A, Nascimento MTC, et al. Neutrophil extracellular trap-enriched supernatants carry microRNAs able to modulate TNF-α production by macrophages. Scientific Reports. 2020;10(1):2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco LP, Pedersen HL, Wang X, Lightfoot YL, Seto N, Carmona-Rivera C, et al. The coenzyme Q10 analog idebenone attenuates murine lupus by improving mitochondrial metabolism and reducing inflammation. Arthritis & Rheumatology.n/a(n/a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka M, Jaruga P, Küpfer PA, Leumann CJ, Dizdaroglu M, Sonntag WE, et al. RNA oxidation catalyzed by cytochrome c leads to its depurination and cross-linking, which may facilitate cytochrome c release from mitochondria. Free Radical Biology and Medicine. 2012;53(4):854–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafner M, Renwick N, Farazi TA, Mihailović A, Pena JTG, Tuschl T. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods. 2012;58(2):164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farazi TA, Brown M, Morozov P, ten Hoeve JJ, Ben-Dov IZ, Hovestadt V, et al. Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods. 2012;58(2):171–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 2010;38(4):576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selth LA, Gilbert C, Svejstrup JQ. RNA Immunoprecipitation to Determine RNA-Protein Associations In Vivo. Cold Spring Harbor Protocols. 2009;2009(6):pdb.prot5234. [DOI] [PubMed] [Google Scholar]
- 22.Goel RR, Wang X, O’Neil LJ, Nakabo S, Hasneen K, Gupta S, et al. Interferon lambda promotes immune dysregulation and tissue inflammation in TLR7-induced lupus. Proceedings of the National Academy of Sciences. 2020;117(10):5409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oulhen N, Foster S, Wray G, Wessel G. Chapter 4 - Identifying gene expression from single cells to single genes. In: Hamdoun A, Foltz KR, editors. Methods in Cell Biology: Academic Press; 2019. p. 127–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahl S, Cerps S, Rippe C, Swärd K, Uller L, Svensson D, et al. Human host defense peptide LL-37 facilitates double-stranded RNA pro-inflammatory signaling through up-regulation of TLR3 expression in vascular smooth muscle cells. Inflammation Research. 2020;69(6):579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrezenmeier E, Dörner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nature Reviews Rheumatology. 2020. [DOI] [PubMed] [Google Scholar]
- 26.Carmona-Rivera C, Carlucci PM, Moore E, Lingampalli N, Uchtenhagen H, James E, et al. Synovial fibroblast-neutrophil interactions promote pathogenic adaptive immunity in rheumatoid arthritis. Science Immunology. 2017;2(10):eaag3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawashima T, Ikari N, Kouchi T, Kowatari Y, Kubota Y, Shimojo N, et al. The molecular mechanism for activating IgA production by Pediococcus acidilactici K15 and the clinical impact in a randomized trial. Scientific Reports. 2018;8(1):5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S-j, Chen Z, Essani AB, Elshabrawy HA, Volin MV, Volkov S, et al. Identification of a Novel Toll-like Receptor 7 Endogenous Ligand in Rheumatoid Arthritis Synovial Fluid That Can Provoke Arthritic Joint Inflammation. Arthritis & Rheumatology. 2016;68(5):1099–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park C-K, Xu Z-Z, Berta T, Han Q, Chen G, Liu X-J, et al. Extracellular microRNAs activate nociceptor neurons to elicit pain via TLR7 and TRPA1. Neuron. 2014;82(1):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Romo GS, Caielli S, Vega B, Connolly J, Allantaz F, Xu Z, et al. Netting Neutrophils Are Major Inducers of Type I IFN Production in Pediatric Systemic Lupus Erythematosus. Science Translational Medicine. 2011;3(73):73ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman LG, Zou J, Crews FT. Microglial-derived miRNA let-7 and HMGB1 contribute to ethanol-induced neurotoxicity via TLR7. Journal of Neuroinflammation. 2017;14(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T-Y, Libardo MDJ, Angeles-Boza AM, Pellois J-P. Membrane Oxidation in Cell Delivery and Cell Killing Applications. ACS Chem Biol. 2017;12(5):1170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ. Neutrophil Extracellular Trap–Associated Protein Activation of the NLRP3 Inflammasome Is Enhanced in Lupus Macrophages. The Journal of Immunology. 2013;190(3):1217–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann A, Berends ETM, Nerlich A, Molhoek EM, Gallo RL, Meerloo T, et al. The antimicrobial peptide LL-37 facilitates the formation of neutrophil extracellular traps. Biochemical Journal. 2014;464(1):3–11. [DOI] [PubMed] [Google Scholar]
- 35.Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, et al. Neutrophils Activate Plasmacytoid Dendritic Cells by Releasing Self-DNA–Peptide Complexes in Systemic Lupus Erythematosus. Science Translational Medicine. 2011;3(73):73ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chamilos G, Gregorio J, Meller S, Lande R, Kontoyiannis DP, Modlin RL, et al. Cytosolic sensing of extracellular self-DNA transported into monocytes by the antimicrobial peptide LL37. Blood. 2012;120(18):3699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salvi V, Gianello V, Busatto S, Bergese P, Andreoli L, D’Oro U, et al. Exosome-delivered microRNAs promote IFN-α secretion by human plasmacytoid DCs via TLR7. JCI Insight. 2018;3(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganguly D, Chamilos G, Lande R, Gregorio J, Meller S, Facchinetti V, et al. Self-RNA–antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. The Journal of Experimental Medicine. 2009;206(9):1983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Raemdonck K, Umar S, Palasiewicz K, Romay B, Volkov S, Arami S, et al. TLR7 endogenous ligands remodel glycolytic macrophages and trigger skin-to-joint crosstalk in psoriatic arthritis. European Journal of Immunology. 2021;51(3):714–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denny MF, Thacker S, Mehta H, Somers EC, Dodick T, Barrat FJ, et al. Interferon-α promotes abnormal vasculogenesis in lupus: a potential pathway for premature atherosclerosis. Blood. 2007;110(8):2907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somers EC, Zhao W, Lewis EE, Wang L, Wing JJ, Sundaram B, et al. Type I Interferons Are Associated with Subclinical Markers of Cardiovascular Disease in a Cohort of Systemic Lupus Erythematosus Patients. PLOS ONE. 2012;7(5):e37000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thacker SG, Berthier CC, Mattinzoli D, Rastaldi MP, Kretzler M, Kaplan MJ. The Detrimental Effects of IFN-α on Vasculogenesis in Lupus Are Mediated by Repression of IL-1 Pathways: Potential Role in Atherogenesis and Renal Vascular Rarefaction. The Journal of Immunology. 2010;185(7):4457–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thacker SG, Zhao W, Smith CK, Luo W, Wang H, Vivekanandan-Giri A, et al. Type I interferons modulate vascular function, repair, thrombosis, and plaque progression in murine models of lupus and atherosclerosis. Arthritis & Rheumatism. 2012;64(9):2975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bao M-h, Zhang Y-w, Lou X-y, Cheng Y, Zhou H-h. Protective Effects of Let-7a and Let-7b on Oxidized Low-Density Lipoprotein Induced Endothelial Cell Injuries. PLOS ONE. 2014;9(9):e106540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beltrami C, Besnier M, Shantikumar S, Shearn AIU, Rajakaruna C, Laftah A, et al. Human Pericardial Fluid Contains Exosomes Enriched with Cardiovascular-Expressed MicroRNAs and Promotes Therapeutic Angiogenesis. Molecular Therapy. 2017;25(3):679–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L, Li Q, Qi D, Niu F, Li Q, Yang H, et al. Atherosclerosis-associated endothelial cell apoptosis by miRNA let7-b-mediated downregulation of HAS-2. Journal of Cellular Biochemistry. 2019;n/a(n/a). [DOI] [PubMed] [Google Scholar]
- 47.Chen P-Y, Qin L, Barnes C, Charisse K, Yi T, Zhang X, et al. FGF Regulates TGF-β Signaling and Endothelial-to-Mesenchymal Transition via Control of let-7 miRNA Expression. Cell Reports. 2012;2(6):1684–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daigo K, Takamatsu Y, Hamakubo T. The Protective Effect against Extracellular Histones Afforded by Long-Pentraxin PTX3 as a Regulator of NETs. Frontiers in Immunology. 2016;7(344). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meegan JE, Yang X, Beard RS, Jannaway M, Chatterjee V, Taylor-Clark TE, et al. Citrullinated histone 3 causes endothelial barrier dysfunction. Biochemical and Biophysical Research Communications. 2018;503(3):1498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saffarzadeh M, Juenemann C, Queisser MA, Lochnit G, Barreto G, Galuska SP, et al. Neutrophil Extracellular Traps Directly Induce Epithelial and Endothelial Cell Death: A Predominant Role of Histones. PLoS ONE. 2012;7(2):e32366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


