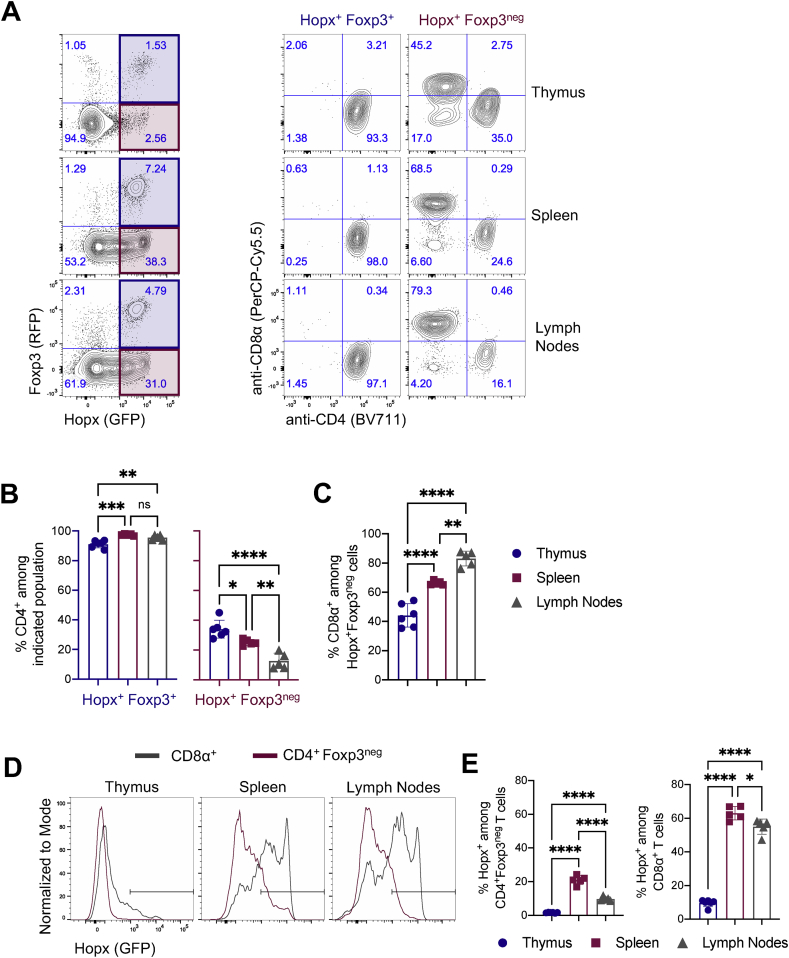
Figure 2.
Multiple T cell subsets express Hopx. Cells from thymi, spleens, and peripheral lymph nodes of HopxGFPFoxp3RFP mice were analyzed by flow cytometry. (A) (Left) Representative plots show Hopx (GFP) and Foxp3 (RFP) expression among all T cells (gated as single, live, CD3ε+, NK1.1neg) from thymus (top), spleen (middle), or lymph nodes (bottom). Shaded regions indicate population further characterized on the right (Hopx+Fopx3+ - blue and Hopx+Foxp3neg – red). (Right) Representative plots show anti-CD4 and anti-CD8α staining intensity among Hopx+Fopx3+ and Hopx+Foxp3neg T cells from the thymus, spleen, and lymph nodes. (B) Graphs show the percentages of CD4+ cells among Hopx+Fopx3+ (left) and Hopx+Foxp3neg (right) T cells from the thymus, spleen, or lymph nodes as indicated (n = 5–6 mice from three independent experiments). (C) Graph shows the percentages of CD8α+ cells among Hopx+Foxp3neg T cells from the thymus, spleen, or lymph nodes as indicated (n = 5–6 mice from three independent experiments). (D) Representative overlaid histograms show Hopx (GFP) expression among CD8α+ (gray) and CD4+Foxp3neg (red) T cells from the thymus, spleen, or lymph nodes as indicated. (E) Graphs show the percentages of Hopx+ cells among CD4+Foxp3neg (left) and CD8α+ (right) T cells from the thymus, spleen, or lymph nodes as indicated (n = 5–6 mice from three independent experiments). (B, C, and E) Graphs show mean ± SD. ns – not significant, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 determined by one-way ANOVA with Tukey's multiple comparisons.