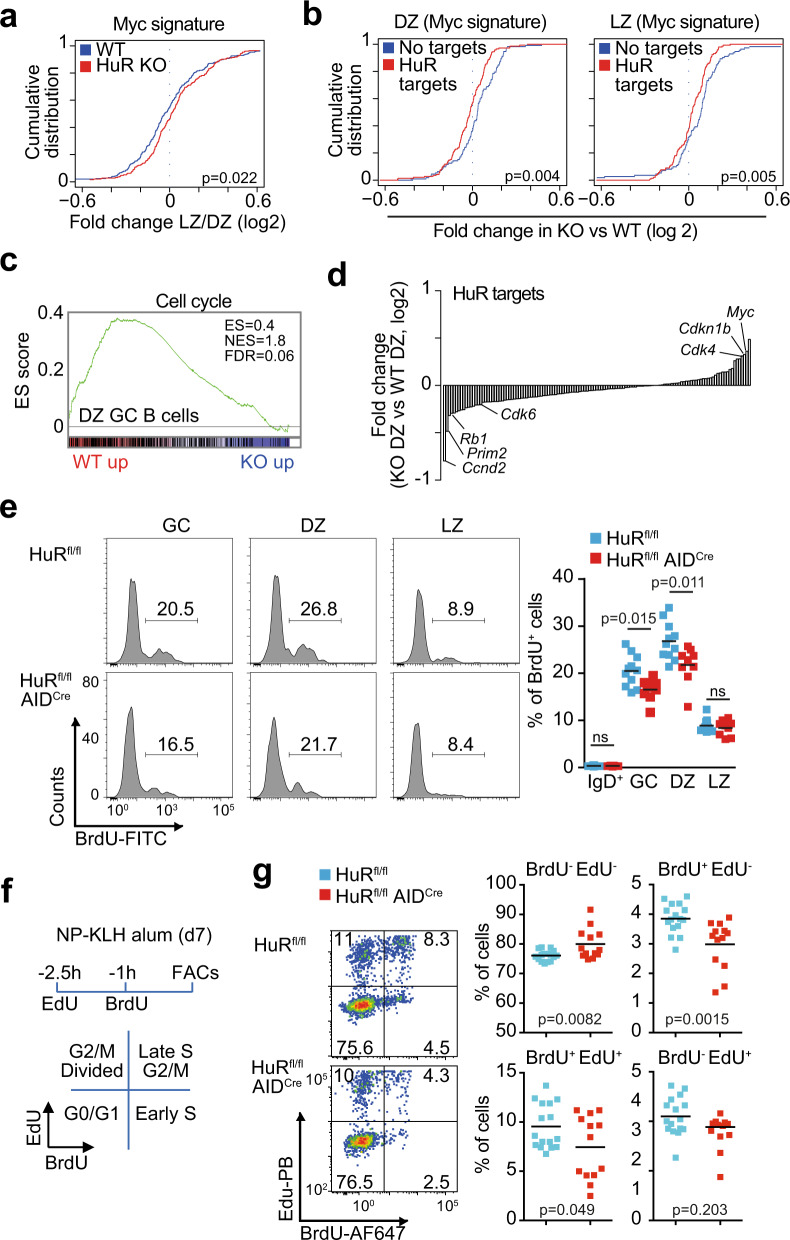
Fig. 7. HuR controls entry and progression through the cell cycle.
a Cumulative distribution of fold change in the expression of Myc-dependent genes (Hallmark gene set) in LZ and DZ GC B cells (Kolmogorov–Smirnov test). b Cumulative distribution of changes in the expression of Myc-dependent genes that are targets or not of HuR (Kolmogorov–Smirnov test). c Enrichment analysis of genes related to cell cycle (GSEA, Gene set from Reactome). d Quantitation from our transcriptome analysis of the changes in the expression of HuR mRNA targets associated with the cell cycle phase G1–S in HuR KO DZ GC B cells compared to control cells (only HuR mRNA targets identified in all conditions of B cells treated with LPS, αCD40+IL4 or αIgM+IL4 were considered). e BrdU incorporation in GC B cells from HuRfl/fl and HuRfl/fl AIDCre mice at day 7 after immunisation with NP-KLH in alum. Left panels, representative histograms of BrdU staining in total GC B cells or in DZ and LZ GC B cells. Right panel, percentage of BrdU+ cells in the cell populations indicated (data from 2 independent experiments, n = 11 (HuRfl/fl) or n = 9 (HuRfl/fl AIDCre) mice, two-sided Mann–Whitney test). f Strategy for labelling proliferating GC B cells in vivo with EdU and BrdU. g Analysis by flow cytometry of cycling GC B cells from HuRfl/fl and HuRfl/fl AIDCre mice at day 7 after immunisation. Left panels, representative contour plots of BrdU and EdU incorporation by cycling GC B cells. Right panels, percentage of cells labelled with EdU, BrdU and EdU/BrdU (data from 2 independent experiments, n = 16 (HuRfl/fl) or n = 13 (HuRfl/fl AIDCre) mice, two-sided Mann-Whitney test). Source data are provided as a Source data file.