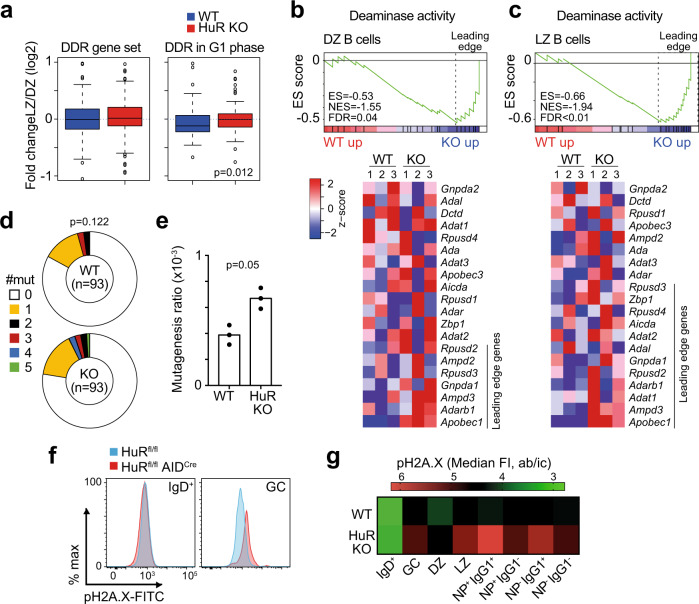
Fig. 8. DNA damage is increased in the absence of HuR.
a Change in the global expression of genes associated with the DNA damage response (n = 119 genes in DDR gene set from Reactome, n = 56 genes in DDR in G1 phase) in LZ and DZ GC B cells (data presented as box plots showing the mean, 25–75% range and whiskers from 5 to 95%, two-sided Wilcoxon test). b, c Enrichment analysis of genes involved in deaminase activity (GSEA, Gene set GO:0019239) in DZ GC B cells (b) and LZ GC B cells (c). Top panels, enrichment plot showing the gene enrichment score (ES), the normalised enrichment score (NES) and the false discovery rate (FDR). Bottom panels, heatmap showing the relative expression of genes involved in deaminase activity in DZ and LZ GC B cells at day 7 after immunisation with NP-KLH in alum. Leading genes driving increased deaminase activity in HuR KO GC B cells are indicated in blue. d Analysis of SHM in GC B cells sorted from HuRfl/fl and HuRfl/fl AIDCre mice at day 7 after immunisation (n = 93 clones analysed per mouse genotype, two-sided Mann–Whitney test). e Quantitation of the mutagenesis ratio in the JH4 intron (n = 3 mice processed independently in different experiments, two-sided Mann–Whitney test). f Flow cytometric histograms showing the expression of pH2A.X (pS139) in naive and GC B cells from HuRfl/fl and HuRfl/fl AIDCre mice at day 14 after immunisation (data representative from 1 of the 2 independent experiments performed, n = 6–9 mice per group). g Heatmap of pH2A.X MFI in the cell types indicated (data is from 2 independent experiments, n = 11–16 mice per group). Source data are provided as a Source data file.