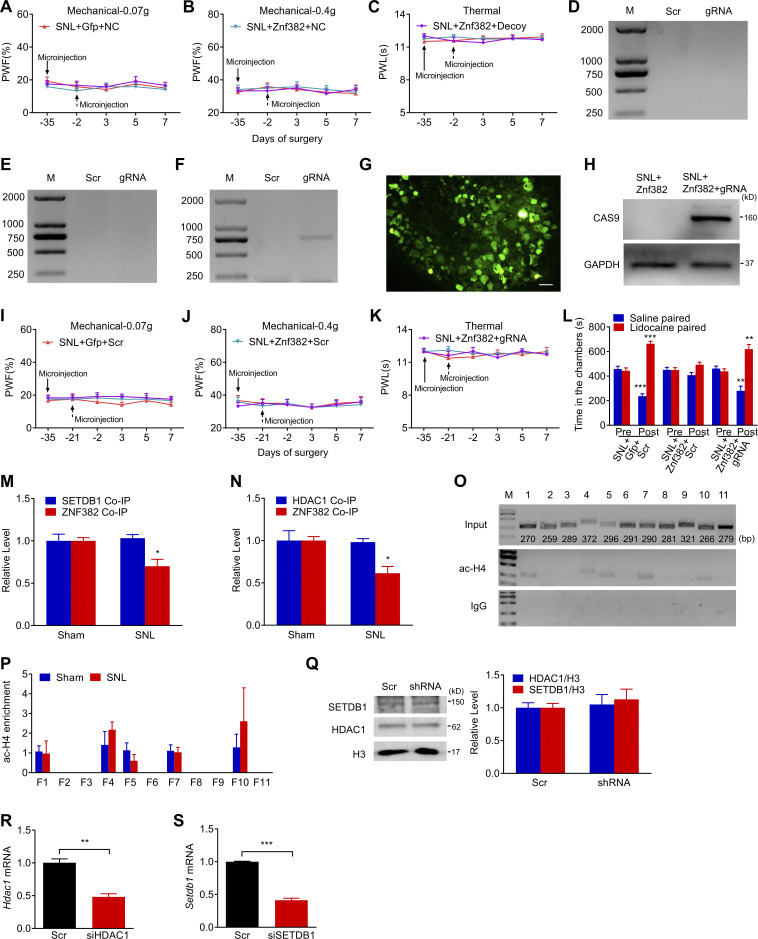
Figure S5.
Validation of LV-gRNA function and effect of decoy DNA or LV-gRNA on basal behavioral responses on the contralateral side. The figure also shows the ac-H4 enrichment in Cxcl13 promoter, validation of HDAC1/SETDB1 knockdown and expression. (A–C) Effect of microinjection with decoy DNA (decoy) or NC (dashed arrow) into the ipsilateral L4 DRG on paw withdrawal responses to mechanical (A and B) and heat (C) stimuli on the contralateral side after SNL surgery from the mice premicroinjected with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. (D–F) PCR-based detection of silencer-included fragment excision of Cxcl13 on day 7 (D), 14 (E), and 21 (F) after LV-gRNA (gRNA) or LV-scrambled gRNA (Scr) microinjection into L4 DRG. n = 2 repeats. Data are representative of two independent experiments. M, ladder marker. (G and H) GFP (G) and CAS9 (H) expression in L4 DRG on day 7 after SNL from the mice premicroinjected with AAV5-Znf382 (Znf382, 35 d before SNL) plus LV-gRNA (gRNA, 21 d before SNL). n = 3 repeats. Data are representative of two independent experiments. Scale bar: 50 μm. (I–K) Effect of microinjection with LV-gRNA (gRNA) or LV-scrambled gRNA (Scr) (dashed arrow) into the ipsilateral L4 DRG on paw withdrawal responses to mechanical (I and J) and heat (K) stimuli on the contralateral side after SNL surgery from the mice premicroinjected with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp). n = 12 mice/group. Data from two independent experiments. Two-way RM ANOVA followed by post hoc Tukey test. (L) Effect of microinjection with LV-gRNA (gRNA) or LV-scrambled gRNA (Scr) into L4 DRG on the time spent in saline- or lidocaine-paired chambers 14 d after SNL surgery from the mice premicroinjected with AAV5-Znf382 (Znf382) or AAV5-Gfp (Gfp). Pre, preconditioning; Post, post-conditioning. n = 9 mice/group. Data from two independent experiments. **, P < 0.01; ***, P < 0.001 versus the corresponding preconditioning by two-way RM ANOVA followed by post hoc Bonferroni’s test. (M and N) ZNF382 and SETDB1 were immunoprecipitated by anti-HDAC1 antibody, and the immunoprecipitated ZNF382 level was decreased after SNL (M), ZNF382 and HDAC1 were immunoprecipitated by anti-SETDB1 antibody, and the immunoprecipitated ZNF382 level was decreased after SNL (N) in the ipsilateral L4 DRG on day 7 after SNL or sham surgery. n = 3 repeats (60 mice)/group. Data from three independent experiments. *, P < 0.05 versus sham group by two-tailed unpaired Student’s t test. (O) Five regions (F1, F4, F5, F7, and F10) from the Cxcl13 promoter and 5′-UTR were immunoprecipitated by ac-H4 antibody in mice L4 DRGs. n = 2 repeats (12 mice). Data are representative of two independent experiments. Input: total purified fragments. M, ladder marker. (P) Level of ac-H4 in F1-F11 from the Cxcl13 promoter in the ipsilateral L4 DRG on day 7 after SNL or sham surgery. n = 3 repeats (18 mice)/group. Data from three independent experiments. Two-tailed unpaired Student’s t test. (Q) Levels of SETDB1 and HDAC1 protein in the ipsilateral L3/L4 DRGs of naive mice 8 wk after AAV5-Znf382 shRNA (shRNA) or AAV5-scrambled shRNA (Scr) microinjection. n = 4 repeats (16 mice)/group. Data from three independent experiments. Two-tailed unpaired Student’s t test. (R and S) Hdac1 mRNA (R) and Setdb1 mRNA (S) knockdown validation in the cultured DRG neurons 3 d after transfection with corresponding Hdac1 siRNA or Setdb1 siRNA. n = 3 repeats. Data from two independent experiments. Two-tailed unpaired Student’s t test. **, P < 0.01; ***, P < 0.001.