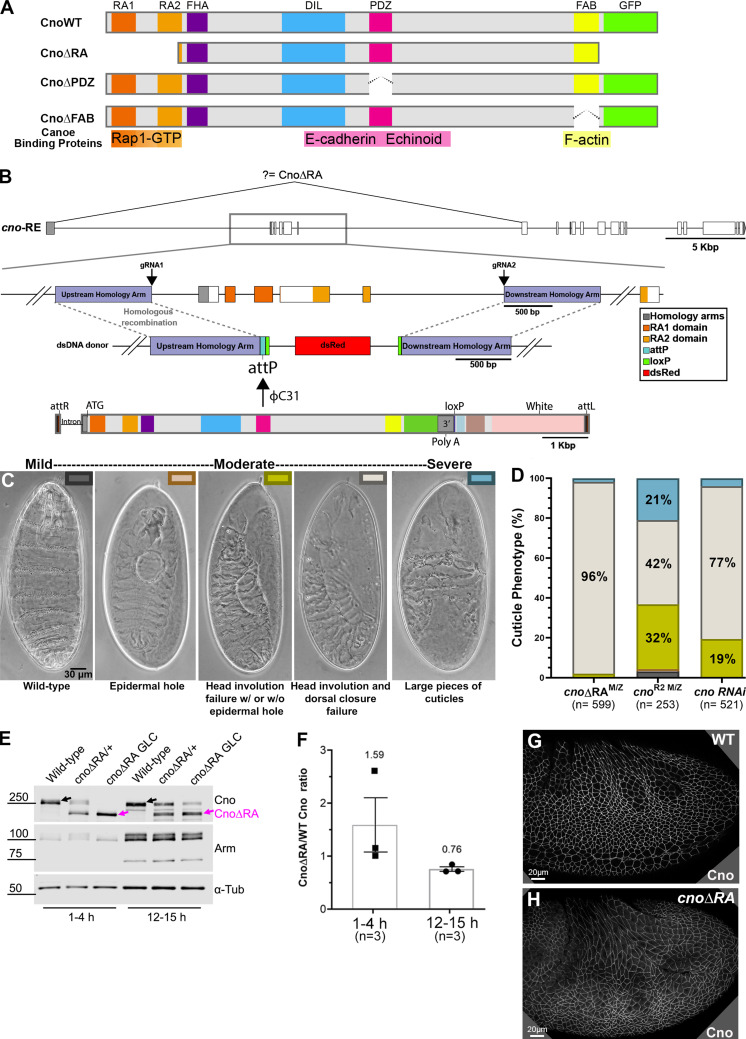
Figure 1.
A platform for site-directed mutants and new insights into RA function. (A) Domain structure of WT Cno and the mutants examined in this work. (B) cno locus. Top: Exons and introns, cno-RE isoform. Middle: Our CRISPR-generated deletion removes exons 2–6 (including start codon) and inserts an attP site and selectable marker (dsRed). Bottom. WT rescue construct. (C) Representative cuticles of cno mutants. (D) Deleting the RA domains substantially reduces Cno function in morphogenesis, leading to failure of head involution and dorsal closure. The data for cnoRNAi and cnoR2 are from Manning et al. (2019). n, total number of cuticles analyzed. (E) Our new mutant produces WT levels of an N-terminally truncated Cno protein. Immunoblot, embryonic extracts, genotypes, and times are indicated (GLC, germline clone). α-Tubulin was used as a loading control. Black arrows indicate WT Cno, and the magenta arrows indicate CnoΔRA. cnoΔRA M/Z mutants exhibit only the truncated protein at early time points (1–4 h), but because 50% of the embryos receive a paternal WT copy of cno, WT Cno protein is seen at the 12–15-h time point. Levels of the AJ protein Arm are not altered in cnoΔRA M/Z mutants. (F) CnoΔRA protein levels relative to WT. Error bars represent SEM, and n is the number of independent replicates. (G and H) After gastrulation onset, CnoΔRA remains localized to AJs.