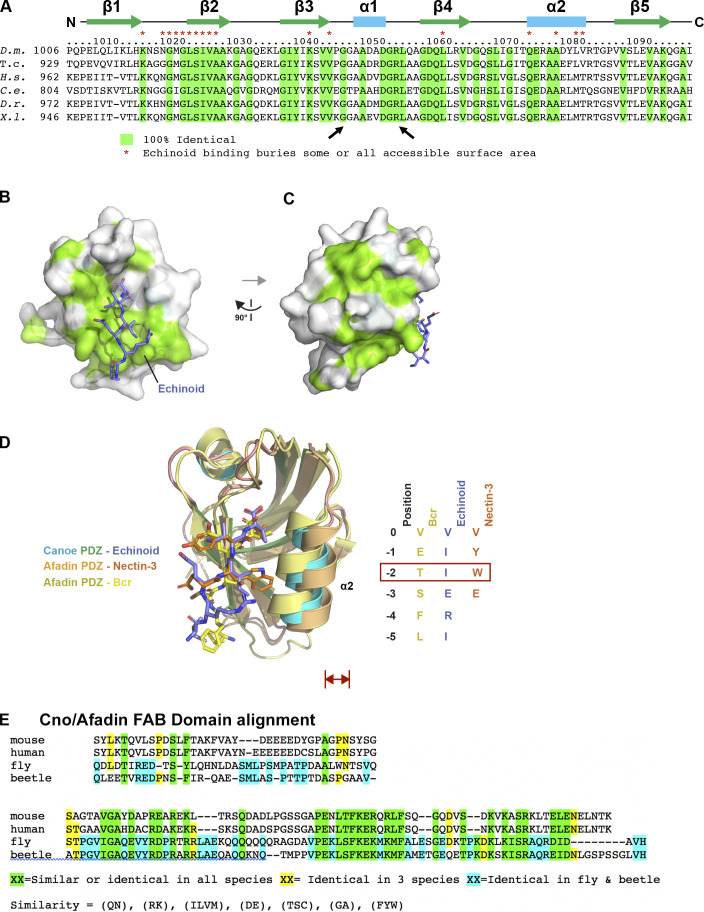
Figure S3.
The Cno PDZ domain has extensive, surface-exposed conservation and displays variation in peptide binding across species. (A) Alignment of Cno/Afadin homologues from Drosophila melanogaster (D.m.), the beetle Tribolium castaneum (T.c.), Homo sapiens (H.s.), Caenorhabditis elegans (C.e.), Danio rerio (D.r.), and Xenopus laevis (X.l.). Residues that are 100% identical across all species are highlighted in green. Cno PDZ domain residue numbering and secondary structure is depicted at top. Cno PDZ domain residues that become partially or totally buried upon Ed binding are denoted above the alignment with a red asterisk. (B) Surface structure of the Cno PDZ domain with conservation (as delineated in A) mapped on the surface. The Ed C-terminal region is shown in stick format, colored purple. The flexible linker was not resolved in protomer A, which is shown in the figure, but it was resolved in protomer B, and the Ed “peptide” that binds protomer A is coming in from protomer B (PDB accession no. 7MFW). (C) Surface structure of the Cno PDZ domain after a 90° rotation of the structure shown in B. (D) Structural alignment of the Cno PDZ–Ed structure (this study), the Afadin PDZ-nectin-3 structure (PDB accession no. 3AXA; Fujiwara et al., 2015), and the AF-6 PDZ-Bcr structure (PDB accession no. 2AIN; Chen et al., 2007). Color-coding each PDZ domain and respective bound peptide is indicated. PDZ domains are shown in ribbon format, and bound peptides are shown in stick format. Peptide sequences of ligands are shown at right, numbered relative to the ultimate valine of each peptide, which is denoted position 0. Residues at position −2 are indicated in a red box, as the size of the side chain at the −2 position correlates with a relative outward shift (red double-headed arrow) of the PDZ domain’s α2 helix. (E) Sequence alignment of the C-terminal regions of Drosophila (fly) and Tribolium (beetle) Cno, and mouse and human Afadin, illustrating the region deleted in CnoΔFAB. The C-terminal 68 aa are highly conserved in insects (81% identity) and share conservation in all homologues (41% similarity). More N-terminal is a region conserved only in insects (39% identity) and not conserved between insects and vertebrates or even between mouse and human Afadin. Sequences immediately more N-terminal are not well conserved among any homologues and begin to include the homopolymeric runs of amino acids found in the intrinsically disordered region.