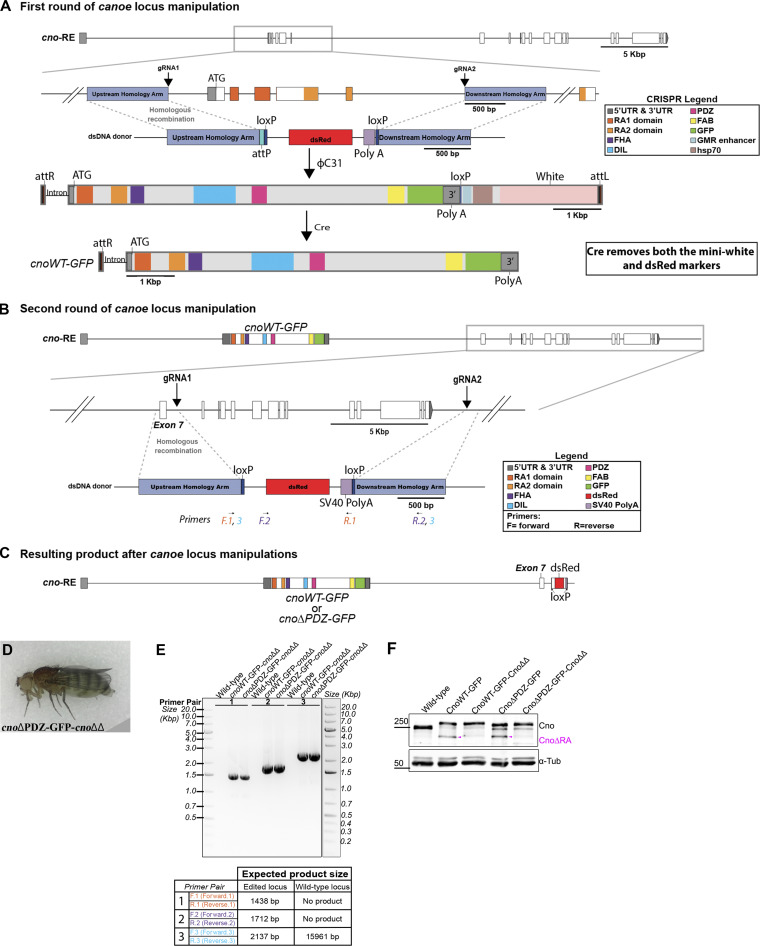
Figure S5.
Platform for deleting residual cnoΔRA sequence in cnoWT-GFP and cnoΔPDZ-GFP mutants. (A) cno locus illustrating the initial ∼3.7-kbp CRIPSR-Cas9 targeted deletion of the translational start site, RA 1 domain and most of RA 2 domain. cnoWT-GFP and cnoΔPDZ-GFP were introduced into the locus using attP site. For the next round of genome manipulation, these were crossed with Cre line to remove both the mini-white and dsRed markers to allow screening for another round of manipulations. (B) Diagram illustrating the second round of cno locus manipulation. Two gRNAs targeting intron sequences deleted almost the rest of the cno coding sequence. gRNA1 targets downstream of exon 7 and gRNA2 targets downstream of 3′ UTR. The deletion was repaired using a donor plasmid with a dsRed marker. (C) Resulting product after multiple manipulations at the cno locus. This generated cnoWT-GFP and cnoΔPDZ-GFP without the residual CnoΔRA protein (cnoΔΔ). (D) Viable homozygous adult female for cnoΔPDZ-GFP-cnoΔΔ. (E) PCR confirmation of the cnoΔΔ deletion in cnoWT-GFP-cnoΔΔ and cnoΔPDZ-GFP-cnoΔΔ mutants using genomic DNA from adult flies. Primers used for the PCR reaction and their expected product size are presented in a table below the agarose gel and in diagram B. Three primers pairs were used to verify the presence of the deletion. Primer pair no. 1 amplifies part of the upstream homology arm through to the SV40 poly(A) signal; this primer pair will only generate a product if the deletion is present. Primer pair no. 2 amplifies from the dsRed marker to downstream of my deletion at the 3′ UTR; this primer set will only generate a product if the deletion is present. Primer pair no. 3 flanks the deletion, and thus both the presence and absence of the deletion can generate a product. However, the product generated by the presence of the deletion is smaller than in WT, and thus, because we are not giving enough time for the WT product to occur, we only obtain a band if the deletion is present. This primer pair would generate a >12-kbp product if the deletion was not present. (F) Immunoblot confirms the absence of the CnoΔRA protein (magenta arrowhead) in both cnoWT-GFP-cnoΔΔ and cnoΔPDZ-GFP-cnoΔΔ mutants. Lysates are embryonic extracts from 1–4 h collection, and α-tubulin was used as a loading control.