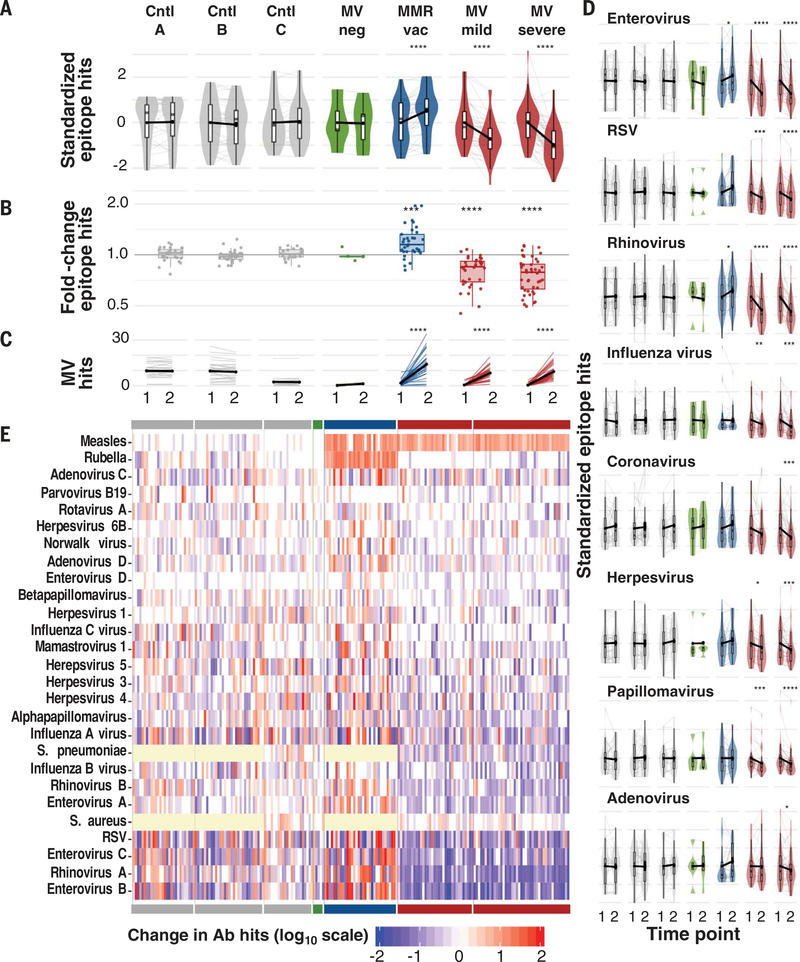
Fig. 1. Measles virus infections reduce antibody diversity.
(A) Total epitopes recognized at time 1, left, and time 2, right, per cohort. For comparison across cohorts, values are standardized across all samples per cohort to a mean of 0 and standard deviation of 1. Each gray line indicates a paired sample from an individual, and black connecting lines indicate mean change from zero. Boxplots indicate interquartile range and median. Asterisks indicate paired t test P values. (B) Fold change of total antibody diversity (i.e., number of total epitope hits) at time 2 versus time 1. Each point represents one paired sample from an individual. Boxplots indicate interquartile range and median. Asterisks indicate significant differences relative to control A (Cntl A), based on student’s t test P values. (C) Number of measles epitope hits per sample at time 1 and time 2. Thin lines indicate paired samples. Black lines indicate cohort averages. (D) As in (A), but for individual viruses. Bonferroni-corrected P values in (A to D): *P < 0.05, **P < 0.001, ***P < 0.001, ****P < 0.0001. (E) Heatmap indicating the change in the total number of epitope hits per species between time 1 and time 2. Each column represents an individual paired sample and each row a pathogen. Cohorts are indicated by the solid bars at top and bottom and are in the same order (left to right) as in (A). Yellow cells indicate that pathogen was not assayed in those samples. Ab, antibody.