Abstract
Microfluidic applications such as active particle sorting or selective enrichment require particle classification techniques that are capable of working in real-time. In this paper, we explore the use of neural networks for fast label-free particle characterization during microfluidic impedance cytometry. A recurrent neural network is designed to process data from a novel impedance chip layout for enabling real-time multi-parametric analysis of the measured impedance data streams. As demonstrated with both synthetic and experimental datasets, the trained network is able to characterize with good accuracy size, velocity and cross-sectional position of beads, red blood cells and yeasts, with a unitary prediction time of 0.4 ms. The proposed approach can be extended to other device designs and cell types for electrical-parameter extraction. This combination of microfluidic impedance cytometry and machine learning can serve as a stepping stone to real time single-cell analysis and sorting.
Keywords: Microfluidic Impedance Cytometry, Multiparametric characterization, Single-cell analysis, Neural Networks, Real-time processing
Graphical Abstract
1. Introduction
Due to their simplicity and label-free nature, electrical sensing techniques are tremendously attractive for the analysis and characterization of particles and cells. Since introduction of the Coulter principle for particle counting [1], the interaction of single particles with electric fields has been exploited in several applications within life sciences, medicine, and environmental monitoring. In particular, in the last two decades, the development of microfabrication technologies has allowed the implementation of microfluidic impedance cytometers for the high-throughput label-free electrical characterization of blood cells, mammalian cell lines, microorganisms and plant cells (see reviews) [2–4].
In a typical microfluidic impedance cytometer, suspended particles flow through a microchannel and integrated electrodes are used to measure the variation in channel impedance induced by the passage of a particle. The impedance change, recorded as an electric current signal, conveys information on geometric and electrical properties of the particle. For instance, depending on the stimulation frequency, signal peak amplitude provides information on particle size, membrane capacitance or intracellular conductivity [5]. Moreover, electrical fingerprints conveying information on both cell intrinsic properties (size, electrical opacity, shape indicators) and cell motion (velocity, cross-sectional position) can be obtained, using particular electrode designs and associated wiring schemes [6–8]. Signal processing of the raw impedance data streams typically involves four steps: preliminary detrending/denoising, data stream segmentation (event detection), extraction of features (like peak amplitude and transit time), and cell classification. Several signal-processing techniques have been implemented in impedance cytometry, like Savitzky-Golay filter for denoising [9,10], wavelet-based or correlation-based approaches for event detection [10,11], least-squares template fitting for feature extraction [7,9], support vector machines or neural networks for classification [12–15]. However, other than protocols based on threshold-based particle counting [16], signal processing of the raw impedance data streams is usually performed post-experiment (offline). Recently, an extended Kalman filtering approach was proposed to develop an impedance-based real-time position sensor [17], and a proof of concept application to determine the longitudinal position of 8.7 μm diameter beads in a channel at a rate of about 2 beads/s was reported. An impedimetric system for real-time monitoring of droplet generation was also presented [18].
For microfluidic applications, such as active particle sorting and selective enrichment [19,20], there is a need for high-throughput real-time signal processing methodologies. Similarly, for distinction of particular cell phenotypes from complex samples with similar sized cells [21], there is a need for routines that can be trained using model cell-types to enable impedance-based signal recognition. Machine learning offers a route to enable automated monitoring of microfluidic systems by converting routinely collected sensor and image data into actionable information in real-time [22,23]. Machine learning has been extensively used for fast image processing in microscopy and flow cytometry applications (see [24–28]). Applications to real-time analysis of unidimensional signals (as opposite to bidimensional images) have also been widely reported in biomedical signal analysis, such as human activity recognition from accelerometer data [29], sleep stage classification from EEG signals [30], patient-specific ECG classification [31], and diabetes detection from breath signals obtained from gas sensors [32]. Recently, Wang et al. [33] used a deep convolutional neural network to analyse code-multiplexed signals from a Coulter sensor network.
In this work, we explore the use of neural networks for real-time feature extraction in microfluidic impedance cytometry. A novel microfluidic impedance chip design for multi-parametric data extraction is presented and the measured signal waveforms are fed to a neural network. Since those signal waveforms are time sequences, a Recurrent Neural Network (RNN) is chosen, which is suited to process data that have a temporal dimension [34,35]. The network is trained to predict cell size, velocity and cross-sectional position (i.e., lateral and vertical) - see Figure 1 for an overview of the proposed system, and Section 2 for details.
Figure 1:
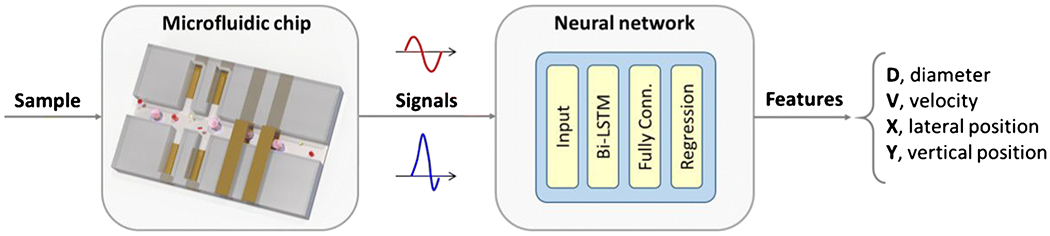
System overview. A suspension of cells flows through the microfluidic impedance cytometer and the electric field perturbation induced by each cell is recorded as a pair of electric current signals. Each signal, representing a high-content cell fingerprint, is fed into a neural network for real-time feature extraction (bi-LSTM, bidirectional Long Short-Term Memory layer; Fully Conn., fully connected layer).
In a preliminary feasibility study, the network is trained and tested on synthetic data streams, and its robustness against measurement noise is investigated. Then, the network is trained and tested on experimentally measured signals from a mixture of polystyrene beads. Finally, cross-sample validation with biologically relevant samples, namely red blood cells (RBCs) and yeast cells suspensions, is performed. The results suggest that this novel approach for impedance pattern recognition can serve as a stepping stone to real-time single-cell analysis and sorting. In particular, rapid determination of cell properties and trajectory can be used to predict cell position under flow conditions for enabling data training and inform downstream sorting based on cellular electrical properties.
2. System Design
2.1. Microfluidic-impedance-chip design
The microfluidic impedance chip considered in this work is shown in Figure 2(a) and (b) (cf. Section 3 for details on chip materials and dimensions). Two subsequent electrical sensing regions are present along the main channel, denoted as H-zone and F-zone, respectively. In the H-zone, two pairs of coplanar electrodes are located at the bottom of lateral channels and the “liquid electrode” concept is exploited to create vertical equipotential surfaces at the apertures of the main channel [36]. Accordingly, an electric field that is uniform along the channel height and non-uniform along channel width (due to fringing effects) is established in the H-zone. In the F-zone, the standard electrode layout comprising two pairs of facing electrodes is used (i.e., the electrodes are located at the top and the bottom of the main channel). This configuration provides an electric field profile that is uniform along channel width and non-uniform along channel height [5].
Figure 2:
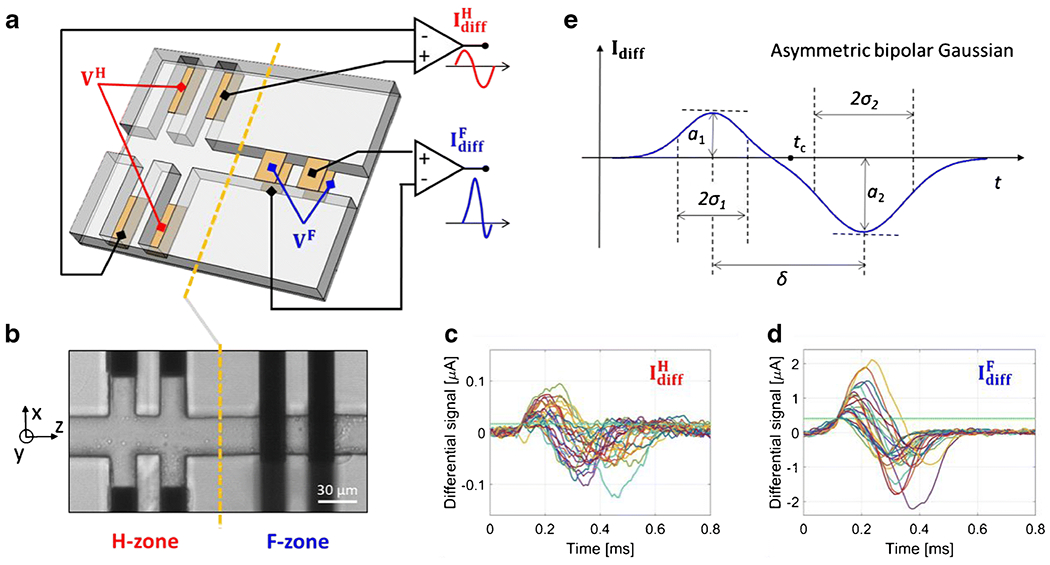
(a) Schematic drawing of the microfluidic impedance chip and of the relevant wiring scheme. Two sensing zones are present, H-zone and F-zone, comprising two pairs of coplanar or facing electrodes, respectively. In each zone, AC voltage is applied to two diagonally opposite electrodes, and the differential current flowing through the remaining two electrodes is collected ( and , respectively). (b) Microscopy image of the microchannel (top view). Dark areas correspond to electrodes. A grounded electrode is locate upstream (not shown). Channel height is 30 μm. The origin of the x and y axes is in the middle of the channel cross-section, (c)-(d) Exemplary event sequences extracted from the signals (c) and (d) . The experimental sample is a mixture of polystyrene beads of 5.2, 6, and 7 μm diameter; twenty-five randomly chosen events are shown in each panel; horizontal green line indicates the threshold used for segmentation, (e) Asymmetric bipolar Gaussian template is used to fit the differential currents and . The fitting parameters a1, a2, and δ allow to estimate particle diameter D, velocity V and cross-sectional position (i.e., X and Y).
Spatial non-uniformity of the electric field is generally regarded as an unwanted feature in microfluidic impedance cytometry, because it introduces positional dependence in the measured electrical signals (i.e., identical particles traveling along different trajectories provide different signals). Unless methods for particle focusing are used, the accuracy of estimated particle properties (e.g., particle size) is reduced due to position blurring. To remove position blurring, we have recently introduced a novel wiring scheme applicable to standard chip layouts involving two electrode pairs (either coplanar or facing) [37,38]. That wiring scheme is used in this work for both the H-zone and the F-zone. In particular, an AC voltage is applied to diagonally opposite electrodes and the differential current flowing through the other two electrodes is collected (Figure 2(a)). When a particle/cell passes through the H and F sensing zones, peculiar differential current signals are recorded. Exemplary event signals, relevant to a mixture of polystyrene beads (5.2, 6 and 7 μm diameter), are reported in Figure 2(c) and (d). For the same applied voltage on the electrodes, the signal-to-noise ratio (SNR) is significantly lower for signal than for signal , due to the larger volume of the sensing region.
The recorded signals have an asymmetric bipolar Gaussian shape (Figure 2(e)), characterized by the following parameters: peak amplitudes (a1 and a2), peak widths (σ1 and σ2), central time (tc) and peak-to-peak time (δ). Those parameters can be extracted from the recorded signals by template fitting. As demonstrated in [37], the peak-amplitude relative difference Δ, defined as:
correlates with particle trajectory. In particular, the peak-amplitude relative difference of the signal from the H-zone () correlates with the lateral position X of the particle (i.e., along channel width); whereas the peak-amplitude relative difference of the signal from the F-zone () correlates with the vertical position Y of the particle (i.e., along channel height). The metric Δ is also used to obtain an electrical estimate of particle diameter D free from position blurring. However, some orientation blurring may affect the estimated size of non-spherical particles (i.e., non-spherical particles traveling with different orientations may provide slightly different signals). Finally, the peak-to-peak time is used to compute particle velocity V. The procedure to obtain electrical estimates of particle diameter, position and velocity from the features of the template is reported in Section 1 of the Electronic Supplementary Material (ESM).
Compared to our previous chip for multiparametric cell characterization [7], the current design has two improvements in data acquisition and extraction. First, the length of the sensing zone is significantly shorter, thereby reducing the fraction of particle coincidence. Second, the approach used to estimate the vertical position is more robust with respect to experimental setting details (cf. Section 2 of the ESM).
2.2. Neural network design
A Recurrent Neural Network (RNN) is designed for fast, possibly real-time, processing of impedance data streams. The RNN is a generalization of the feedforward neural network that has an internal memory, and therefore it is suited to process data and sequences that have a temporal dimension [34,35]. RNNs have been used, for instance, to recognize patterns in sequences of data, such as text and handwriting, or numerical time series data emanating from sensors. In particular, a Long Short-Term Memory (LSTM) network is considered here, which is a modified version of the introduced RNN, in order to mitigate the vanishing gradient problem [39,40]..
The network architecture is composed of four layers (Figure 1): an input layer, a bidirectional Long Short-Term Memory (bi-LSTM) layer, a fully connected layer, and a regression layer. The input layer receives as input the differential current signal ( or ) measured when a particle/cell is passing through the sensing region (H-zone or F-zone, respectively). Identification of the corresponding portions of the data stream (i.e., event detection) is performed by means of thresholding on signal amplitude. The length of the input sequences is chosen on the basis of channel geometry and flow rate, in such a way that the longest signals (i.e., slowest particles) are entirely captured.
The bi-LSTM layer has one hundred hidden units and uses a hyperbolic tangent activation function. The output of the bi-LSTM layer feeds a fully connected layer with three outputs, one for each feature (i.e., diameter, X or Y position, velocity). Finally, a regression layer computes the half-mean-squared-error of the predicted responses. The electrical estimates of particle properties (namely, D, X or Y, and V) obtained via feature extraction based on template-fitting are used as target features in the training, validation and testing phases. Those features are normalized in the range [0,1]. The predictions of the neural network are then scaled back to their original ranges in the testing phase.
Although it would be possible to design a neural network that receives as input both signals and and provides four outputs (diameter, velocity, X-position and Y-position), two distinct networks are considered in this work, because the two signals have significantly different SNR.
3. Materials and Methods
The chip was fabricated by patterning 20 nm Ti/ 200 nm Au electrodes on two 4” diameter glass wafers (D263, Schott) by e-beam evaporation (CHA-50, CHA). Electrode width along channel length is 20 μm, with a 20 μm spacing between the electrode pairs of each sensing zone. Electrodes in lateral channels are recessed by 30 μm from the main channel. The center-to-center distance of the H-zone and F-zone is 120 μm. The microchannel design (30 μm x 30 μm cross-section) was fabricated on one wafer using SU8. The substrates were aligned (mask aligner EVG620, EV Group) and thermally bonded. Individual dies were then cut using a dicing saw (DAD3220, Disco) and fluidic access points laser-drilled. A custom holder was used to connect the microfluidic chip with the electronic acquisition system and fluidic pump (cf. Fig. S2 of the ESM).
The differential currents and were measured using two transimpedance amplifiers (HF2TA, Zurich Instruments) and an impedance spectroscope (HF2IS, Zurich Instruments, working at 115 kSa/s sampling rate, 30 kHz filter bandwidth). An excitation signal VH at 615 kHz was used in the H-zone (i.e., for acquiring ), and an excitation signal VF at 500 kHz was used in the H-zone (i.e., for acquiring ), in order to minimize interference between the two channels.
Samples comprising a mixture of beads (5, 6 and 7 μm nominal diameters; 5.2, 6 and 7 μm lot mean diameters) were used to obtain impedance data for network training, validation and testing. In order to perform cross-sample validation, data relevant to red blood cells and yeast cells suspensions were also acquired. Blood was collected by a finger prick from a healthy donor in accordance with ethical standards of the protocol approved by the Institutional Review Board (see section on “Compliance with Ethical Standards”). Biological samples were spiked with 7 or 6 μm diameter beads, respectively, for internal reference. In fact, at the stimulation frequencies used herein, biological particles behave like insulating particles [5]. In all experiments, typical sample concentrations were around 106 particles/ml. A syringe pump (Elite 11, Harvard Apparatus) working at 10 μl/min was used for fluidic control. Representative images of flowing particles/cells are provided in Figs. S3 and S4 of the ESM. In particular, RBCs exhibited a normal shape (biconcave disc) and were traveling with different orientations, e.g. edge-on (ESM Fig. S4, panels a-c) or face-on (ESM Fig. S4, panels d-e).
The measured impedance data streams were processed with a tailored toolbox developed in Matlab. All computations were run on a Intel Xeon CPU E5-2660 v3 @2.60 GHz processor with 128 GB RAM.
In addition, a finite element model of the microfluidic impedance chip (a so-called virtual device) was developed and used to provide close-to-real synthetic data streams. The latter were used to test the feasibility of the proposed neural-network-based approach, in a preliminary investigation phase. Details on how to build a virtual device are reported in Caselli and Bisegna [41]. Finite element simulations were also used to assess the effect of possible misalignment between the top and bottom electrodes of the F-zone (cf. Fig. S5 of the ESM).
4. Results and discussion
4.1. Multiparametric Impedance-based characterization
The differential current signals and convey high-content information. As detailed in Section 2.1, feature extraction based on template fitting and simple calibration provide electrical estimates of individual particle size, velocity and cross-sectional position within the microchannel. In particular, each signal ( or ) provides three features: diameter, velocity and position (X or Y, respectively). Diameter and velocity estimates obtained from are more accurate than those obtained from , due to the higher SNR of the former signal (cf. Fig. S6 of the ESM).
As an example, Figure 3 shows experimental results relevant to the multiparametric characterization of a mixture of polystyrene beads (5.2, 6, and 7 μm diameter). The density plots of particle cross-sectional distribution (Figure 3(a)) and velocity profile (Figure 3(b)) enable an optics-free system for monitoring of particle motion. The histogram of particle diameter (Figure 3(c)) shows the capability of the system to provide accurate particle sizing. The scatter plots of particle cross-sectional distribution and velocity profile labelled according to the bead population are reported in Fig. S7 of the ESM.
Figure 3:
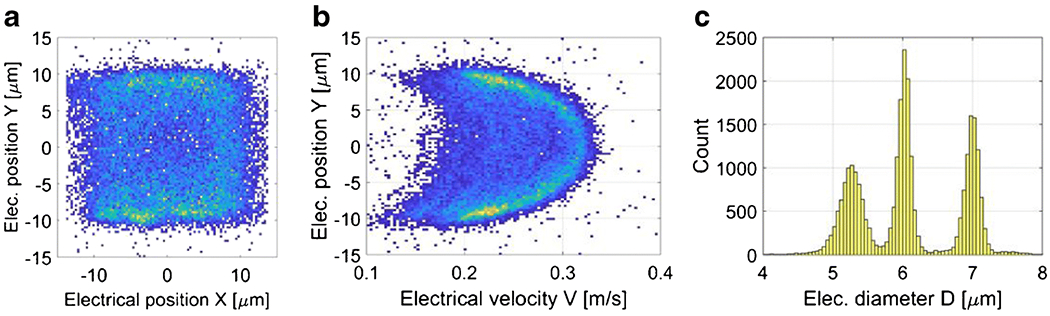
Multiparametric characterization of a mixture of polystyrene beads (5.2, 6, and 7 μm diameter). (a) Density plot of particle distribution in the channel cross-section. (b) Density plot of particle velocity against particle vertical position. (c) Histogram of particle diameter. Velocity and diameter were obtained from the signal . About 27000 events are reported.
4.2. Training, validation and testing on synthetic data streams
As a preliminary feasibility study, the network was trained and subsequently tested on virtual data with different levels of noise (i.e., SNR in the range 30-6.5 dB). In particular, synthetic data streams relevant to a mixture of beads (5, 6, and 7 μm diameter) measured in the F-zone were considered. Simulation parameters were tuned to mimic the experimental conditions of the actual experiments (e.g., in terms of sample concentration, flow-rate, and properties of the electronic acquisition system). Exemplary synthetic traces are reported in Fig. S8 of the ESM.
The network was trained on approximately 23000 events from the dataset with highest SNR (30 dB). The electrical features obtained by means of template fitting were used as targets for the supervised training of the network. The total time required to train the network was 39 min. The trained network, referred to as Net-S, was tested on all datasets. Figure 4(a)-(c) reports the testing results relevant to the data stream with SNR 30 dB. In particular, the density plots of predicted features versus target features are shown for particle diameter (panel a), particle velocity (panel b) and particle position (panel c). The root mean squared error (RMSE) of the predicted values with respect to the target values are reported in Table 1. A RMSE of 0.12 μm is found for particle diameter. RMSE on velocity is 1.8% of the maximum velocity, whereas RMSE on position is 2.2% of channel width. RMSE values obtained when testing the network Net-S on data streams with lower SNR (i.e., 24, 16, 10, or 6.5 dB) are also reported in Table 1, showing that the proposed approach is reasonably robust with respect to measurement noise. In all tests, the prediction time for single event was about 0.4 ms (cf. Unitary time in Table 1).
Figure 4:
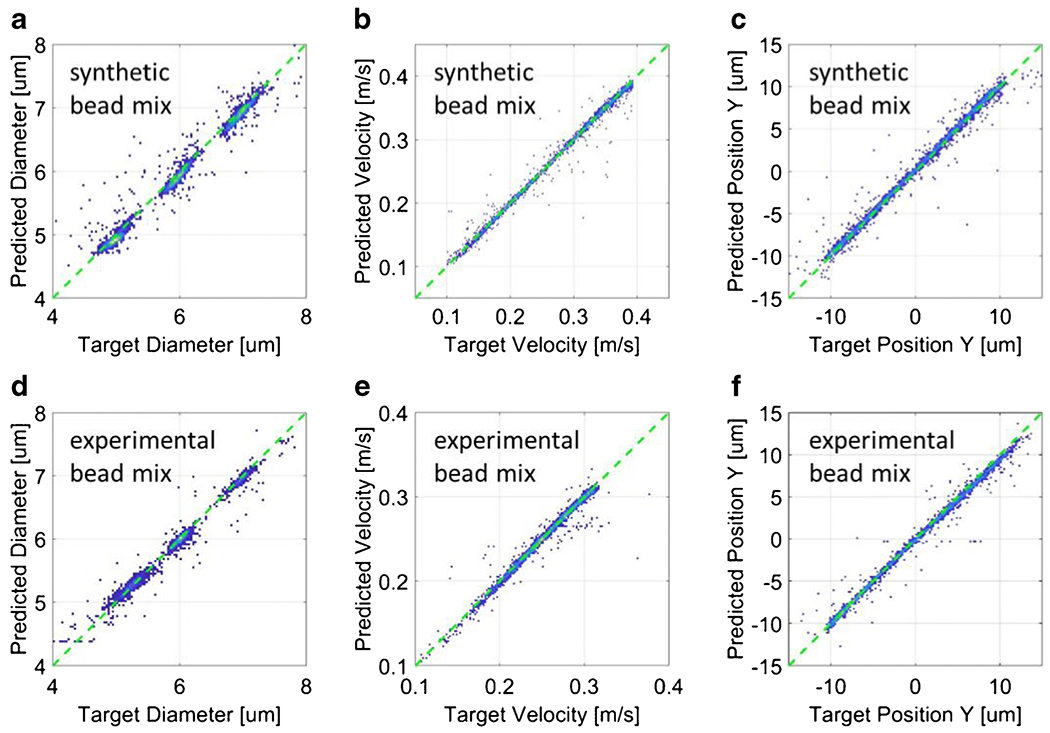
(a)-(c) Neural network Net-S tested on a synthetic mixture of beads (5, 6 and 7 μm diameter; SNR=30 dB). (d)-(f) Neural network Net-1 tested on an experimental mixture of beads (5.2, 6 and 7 μm diameter). Density plots of predicted features against target features: (a) and (d) particle diameter, (b) and (e) particle velocity, (c) and (f) particle vertical position. Root mean squared error values are reported in Table 1.
Table 1:
Summary of testing results on synthetic or experimental datasets. For each test, the unitary prediction time and the root mean squared error (RMSE) of predicted values with respect to target values are reported.
| Testing dataset | Network | Number of events | Unitary time [ms] | RMSE diameter [μm] | RMSE velocity [%] | RMSE position [%] |
|---|---|---|---|---|---|---|
| Synthetic data | ||||||
| Bead mix @ 30 dB SNR | Net-S | 4000 | 0.42 | 0.12 | 1.8 | 2.2 |
| Bead mix @ 24 dB SNR | Net-S | 16000 | 0.39 | 0.12 | 1.9 | 2.3 |
| Bead mix @ 16 dB SNR | Net-S | 16000 | 0.42 | 0.14 | 2.2 | 2.7 |
| Bead mix @ 10 dB SNR | Net-S | 16000 | 0.41 | 0.22 | 4.3 | 4.2 |
| Bead mix @ 6.5 dB SNR | Net-S | 16000 | 0.40 | 0.27 | 5.0 | 4.8 |
| Experimental data | ||||||
| Bead mix | Net-1 | 2570 | 0.46 | 0.09 | 2.2 | 2.4 |
| RBCs + 7 μm beads | Net-1 | 41338 | 0.40 | 0.14 | 3.2 | 4.1 |
| Yeasts + 6 μm beads | Net-1 | 24613 | 0.41 | 0.25 | 7.3 | 11.5 |
| Bead mix, RBCs, Yeasts | Net-2 | 12388 | 0.41 | 0.16 | 2.4 | 2.9 |
4.3. Training, validation and testing on experimental data streams
After the preliminary investigation on synthetic data streams, the neural network was trained on experimental signals from a mixture of 5.2, 6 and 7 μm diameter polystyrene beads. The complete dataset comprised approximately 17000 single-particle signals and was divided into three subgroups of data for training (70%), validation (15%), and testing (15%). The electrical features obtained by means of template fitting were used as targets for the supervised training of the neural network. The total time required to train the network was 25 min. The trained network is hereafter denoted as Net-1.
Testing results are reported in Figure 4(d)-(f). The density plots of predicted features versus target features show an excellent correlation, with RMSE values of 0.09 μm, 2.2%, and 2.4%, for particle diameter, velocity and position, respectively (cf. Table 1). The overlapping histograms of target features and predicted features are also reported (Figure 5(a)-(c)). In the histogram of particle diameter, the three bead populations are clearly distinguishable and follow Gaussian distributions. This confirms that position-induced blurring was effectively removed. The histograms of particle velocity and particle position show a non-uniform distribution of particles across the channel cross-section, with a focusing region in the lower half of the channel, possibly due to the combined effects of sedimentation and inertia.
Figure 5:
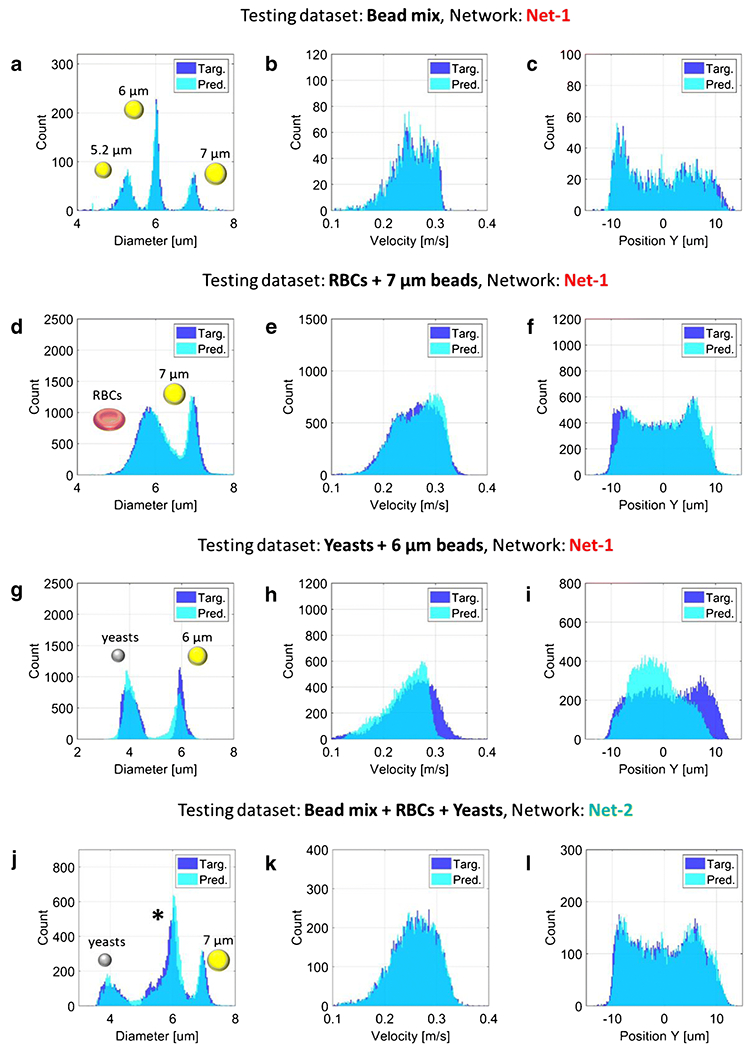
(a)-(i) Testing of the neural network Net-1 on: (a)-(c) a mixture of beads (5.2, 6 and 7 μm diameter), (d)-(f) red blood cells spiked with 7 μm beads, and (g)-(i) yeasts cells spiked with 6 μm beads, (j)-(l) Testing of the neural network Net-2 on a mixture of beads, red blood cells and yeast cells. In panel (j) the central population, marked with an asterisk, comprises 5.2 μm and 6 μm diameter beads and RBCs. The latter could be distinguished from beads by means of electrical opacity (e.g., [7]). The histograms of diameter, velocity and vertical position are reported in the first, second and third column, respectively. RBC diameter indicates the diameter of a sphere with the same volume. Root mean squared error values of predicted versus target features are reported in Table 1.
Analogous results relevant to the signal are reported in Fig. S9 of the ESM.
The network Net-1 (trained on the experimental bead mixture) was also tested on experimental datasets relevant to measurements of red blood cells or yeasts suspensions. This investigation was aimed at assessing the applicability of the approach to biologically relevant samples, as well as evaluating its robustness with respect to sample type. The results are shown in Figure 5(d)-(f) for the RBCs, and Figure 5(g)-(i) for the yeasts, and the RMSE values of the predicted features are reported in Table 1. While good results are obtained for the RBCs, less satisfactory results are obtained for the yeasts. This can be attributed to the fact that the yeasts are smaller in size than the particle used in the training dataset. In addition, their reduced size implies a reduced SNR. In order to improve prediction quality on yeasts, the network was trained on a dataset comprising a mixture of beads, RBCs and yeast cells. The total time required to train this network, referred to as Net-2, was 112 min. The testing results are reported in Figure 5(j)-(l), and show excellent performances (cf. also Table 1, last row).
In all the experiments reported in Table 1, the time required by the neural networks t6o predict the feature of a single-event (unitary time) was about 0.4 ms. By comparison, an optimized feature extraction based on template-fitting required a unitary time of about 3 ms. Accordingly, the proposed neural-network-based approach enables 7.5-fold enhancement in data processing speed. While microfluidic actuation systems with response times in the ~10 ms timeframe have been developed [42], the signal recognition step remains a bottleneck [17]. Hence, the current study addresses the pressing need for simple and label-free real-time recognition approaches.
The quality of neural network predictions greatly depends on the quality of the training dataset, which should be as rich and complete as possible. In microfluidic impedance cytometry, dealing with unidimensional signals, the acquisition and storage of a large number of examples to include in the training dataset is less demanding than in image processing applications. Data augmentation strategies can also be implemented [43], like stretching the impedance signals to mimic different flow-rates or using synthetic data obtained via numerical simulations.
Training of the neural network is based on supervised learning, which means that the training dataset has to be labelled. Similarly, benchmark values are needed for testing the network performance. In this work, the features (diameter, velocity and position) computed by means of least-square template-fitting on impedance signals have been used as labels (i.e., target values). Their accuracy has been assessed in our previous works [7,37,44]. Alternatively, features computed by means of image processing could be used [33]. However, the latter approach requires high-speed image acquisition systems and the estimation of cell properties is not always straightforward, due to possible motion blurring, low contrast and limited spatial resolution. Moreover, estimating particle vertical position from projection images may require the implementation of a quantitative defocusing approach [45].
To increase the richness of cell characterization, electric phase information or higher stimulation frequencies can be considered, for instance, to retrieve information related to cell viability, plasma membrane morphology or intracellular properties [21,46–48]. Moreover, additional sensing zones can be introduced able to provide cell shape indicators [8,49].
The approach proposed in this work can be used to develop new label-free cell sorting techniques for identification and sample pre-enrichment. As an example, a system tailored for detection and separation of yeast cells based on their geometric or electrical characteristics could be developed [49,50], which is critical to the investigation of cell division cycles [51]. As another example, since electrical fingerprints correlates with the stages of Plasmodium falciparum infection of red blood cells [52], sorting strategies aimed at the concentration of parasitized cells within the blood sample could be devised. The enriched sample could then be detected using current diagnostic methods with enhanced sensitivity, which is crucial for malaria diagnosis in asymptomatic individuals with very low parasite densities.
Conclusions
In this work, an innovative machine learning approach for fast signal processing in microfluidic impedance cytometry was presented. A recurrent neural network was designed to predict cell diameter, velocity and position from electric current signals measured with a novel microfluidic impedance chip. The feasibility of the approach was tested on both synthetic data streams and experimental ones. The network was able to characterize with good accuracy geometric and electrical properties of beads, red blood cells and yeasts. The average unitary prediction time was 0.4 ms, which amounts to a theoretical rate of 2500 cells/s.
The results demonstrate that the combination of microfluidic impedance cytometry and machine learning enables the label-free multiparametric characterization of biological cells at high-throughput potentially in real-time. This paves the way for the effective application of microfluidic impedance cytometry in diagnostics, life science, and personalized medicine.
Supplementary Material
Funding Sources:
Funding from the Italian Ministry of Education, University and Research (SIR 2014, Grant RBSI14TX20), from the University of Rome Tor Vergata (Mission Sustainability, Grant E81I18000540005), from the U.S. National Center for Advancing Translational Sciences of the National Institutes of Health (Award Number UL1TR003015), and from Advanced Regenerative Medicine Institute’s BioFab-USA (Subcontract T0163) are acknowledged.
Biographies

Carlos Honrado received his Ph.D. from the School of Electronics and Computer Science at the University of Southampton (UK) and he is now further pursuing research on label-free single particle analysis and separation, as a postdoctoral research associate position in the Department of Electrical and Computer Engineering at the University of Virginia (USA). His research interests are focused on the development of microfluidic devices for biomedical applications, using label-free and single-cell methods based on AC electrokinetics. He is a member of the Biomedical Engineering Society (BMES).

John S. McGrath received an undergraduate in Cell Biology and PhD in Biotechnology at Heriot-Watt University (UK), the latter of which was focused on developing microfluidic tools for sorting and/ or detecting waterborne pathogens. He then held a postdoctoral position at the University of Virginia (USA), during which he designed and fabricated microfluidic impedance cytometers for use in biomedical applications. John is now a senior researcher at the University of Glasgow (UK) and a scientific project manager at EVOdrops ITN. His research interests include high-throughput microfluidic screening and/ or sorting techniques, droplet microfluidics, acoustofluidics, microfluidic integration, and microfabrication.

Riccardo Reale is a researcher in the department of Civil Engineering and Computer Science at the University of Rome Tor Vergata. He received his PhD from the University of Southampton, UK. His research interests are focused on the development of innovative lab-on-chip devices for cellular analysis (e.g. organs-on-chip, microfluidic impedance cytometer).

Paolo Bisegna serves as Professor of Mechanics of Materials and Structures at the Department of Civil Engineering and Computer Science, University of Rome Tor Vergata, and as Director of the Structural Engineering Doctoral Program. He received the M.Sc. degree in Engineering, the M.Sc. degree in Mathematics, and the M.D. degree. His research interests include lab-on-a-chip devices, biomechanics, mechanics of materials and structures.

Nathan S. Swami serves as Professor of Electrical & Computer Engineering at the University of Virginia (UVA), Charlottesville, VA. His research group specializes in label-free microfluidic techniques for biofabrication, electrophysiology-based single-cell analysis and nano-confined systems for biomolecular analysis. Previously, he served on the scientific staff of the MEMS group at Motorola Labs and at Clinical Microsensors, Inc. He seeks to impact diagnostic systems within point-of-care and resource-poor settings for precision medicine.

Federica Caselli is Associate Professor in Biomedical Engineering at University of Rome Tor Vergata. She graduated in Medical Engineering, in Mathematics, and obtained her Ph.D. in advanced computational methods in biomechanics. Dr. Caselli’s research deals with the development of lab-on-a-chip devices for diagnostics and life science, with a special focus on microfluidic impedance cytometry for single-cell biophysical phenotyping. She has published about a hundred contributions in journals, conference proceedings and book chapters, and she was awarded a grant from the “Scientific Independence of Young Researchers Programme”.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Compliance with Ethical Standards: The reported studies on blood samples have been approved by the University of Virginia Institutional Review Board for Health Sciences Research (IRB-HSR protocol #21081) and have been performed in accordance with ethical standards.
Conflicts of Interest Declaration: The authors have no conflicts of interest on the reported material. The reported content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
References
- 1.Coulter WH. Means for counting particles suspended in a fluid. US 2656508 A, 1953.
- 2.Cheung KC, Di Berardino M, Schade-Kampmann G, Hebeisen M, Pierzchalski A, Bocsi J, Mittag A, Tárnok A. Microfluidic impedance-based flow cytometry. Cytom Part A. 2010;77(7):648–66. [DOI] [PubMed] [Google Scholar]
- 3.Petchakup C, Li KHH, Hou HW. Advances in Single Cell Impedance Cytometry for Biomedical Applications. Micromachines. 2017;8(3). [Google Scholar]
- 4.Vembadi A, Menachery A, Qasaimeh MA. Cell cytometry: review and perspective on biotechnological advances. Front Bioeng Biotechnol [Internet]. 2019;7:147. Available from: 10.3389/fbioe.2019.00147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gawad S, Cheung K, Seger U, Bertsch A, Renaud P. Dielectric spectroscopy in a micromachined flow cytometer: theoretical and practical considerations. Lab Chip. 2004;4(3):241–51. [DOI] [PubMed] [Google Scholar]
- 6.Reale R, Ninno A De, Businaro L, Bisegna P, Caselli F. Electrical measurement of cross - sectional position of particles flowing through a microchannel. Microfluid Nanofluid [Internet]. 2018;22(41):1–13. Available from: 10.1007/s10404-018-2055-3 [DOI] [Google Scholar]
- 7.Reale R, De Ninno A, Businaro L, Bisegna P, Caselli F. High-throughput electrical position detection of single flowing particles/cells with non-spherical shape. Lab Chip [Internet]. 2019;19(10):1818–27. Available from: 10.1039/C9LC00071B [DOI] [PubMed] [Google Scholar]
- 8.McGrath J, Reale R, Honrado C, Bisegna P, Swami N, Caselli F. Towards real-time multiparametric impedance cytometry. In: 23nd International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS 2019). 2019. [Google Scholar]
- 9.Sun T, van Berkel C, Green NG, Morgan H. Digital signal processing methods for impedance microfluidic cytometry. Microfluid Nanofluid. 2009;6(2):179–87. [Google Scholar]
- 10.Evander M, Ricco AJ, Morser J, Kovacs GTA, Leung LLK, Giovangrandi L. Microfluidic impedance cytometer for platelet analysis. Lab Chip. 2013;13(4):722–9. [DOI] [PubMed] [Google Scholar]
- 11.Caselli F, Bisegna P. A simple and robust event-detection algorithm for single-cell impedance cytometry. IEEE Trans Biomed Eng. 2016;63(2):415–22. [DOI] [PubMed] [Google Scholar]
- 12.Guo J, Chen Z, Ban Y, Kang Y. Precise enumeration of circulating tumor cells using support vector machine algorithm on a microfluidic sensor. IEEE Trans Emerg Top Comput. 2014;5(4):518–25. [Google Scholar]
- 13.Ahuja K, Rather GM, Lin Z, Sui J, Xie P, Le T, Bertino JR, Javanmard M. Toward point-of-care assessment of patient response: a portable tool for rapidly assessing cancer drug efficacy using multifrequency impedance cytometry and supervised machine learning. Microsyst Nanoeng [Internet]. 2019;5(1):34. Available from: 10.1038/s41378-019-0073-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Xue C, Zhao Y, Chen D, Wu MH, Wang J. Microfluidic impedance flow cytometry enabling high-throughput single-cell electrical property characterization. Int J Mol Sci. 2015;16(5):9804–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Wang K, Chen D, Fan B, Xu Y, Ye Y, Wang J, Chen J, Huang C. Development of microfluidic impedance cytometry enabling the quantification of specific membrane capacitance and cytoplasm conductivity from 100,000 single cells. Biosens Bioelectron [Internet]. 2018;111:138–43. Available from: http://www.sciencedirect.com/science/article/pii/S0956566318302756 [DOI] [PubMed] [Google Scholar]
- 16.Furniturewalla A, Chan M, Sui J, Ahuja K, Javanmard M. Fully integrated wearable impedance cytometry platform on flexible circuit board with online smartphone readout. Microsyst Nanoeng [Internet]. 2018;4(1):20. Available from: 10.1038/s41378-018-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brazey B, Cottet J, Bolopion A, Van Lintel H, Renaud P, Gauthier M. Impedance-based real-time position sensor for lab-on-a-chip devices. Lab Chip [Internet]. 2018;18(5):818–31. Available from: 10.1039/C7LC01344B [DOI] [PubMed] [Google Scholar]
- 18.Saateh A, Kalantarifard A, Celik OT, Asghari M, Serhatlioglu M, Elbuken C. Real-time impedimetric droplet measurement (iDM). Lab Chip [Internet]. 2019;19(22):3815–24. Available from: 10.1039/C9LC00641A [DOI] [PubMed] [Google Scholar]
- 19.Farmehini V, Varhue W, Salahi A, Hyler AR, Čemažar J, Davalos R, Swami NS. On-Chip Impedance for Quantifying Parasitic Voltages During AC Electrokinetic Trapping. IEEE Trans Biomed Eng. 2019;1. [DOI] [PubMed] [Google Scholar]
- 20.Rohani A, Sanghavi BJ, Salahi A, Liao K-TT, Chou C-FF, Swami NS. Frequency-selective electrokinetic enrichment of biomolecules in physiological media based on electrical double-layer polarization. Nanoscale [Internet]. 2017;9(33):12124–31. Available from: 10.1039/C7NR02376F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrath JS, Honrado C, Moore JH, Adair SJ, Varhue WB, Salahi A, Farmehini V, Goudreau BJ, Nagdas S, Blais EM, Bauer TW, Swami NS. Electrophysiology-based stratification of pancreatic tumorigenicity by label-free single-cell impedance cytometry. Anal Chim Acta [Internet]. 2019; Available from: http://www.sciencedirect.com/science/article/pii/S0003267019314898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riordon J, Sovilj D, Sanner S, Sinton D, Young EWK. Deep Learning with Microfluidics for Biotechnology. Trends Biotechnol [Internet]. 2019;37(3):310–24. Available from: http://www.sciencedirect.com/science/article/pii/S0167779918302452 [DOI] [PubMed] [Google Scholar]
- 23.Chu A, Nguyen D, Talathi SS, Wilson AC, Ye C, Smith WL, Kaplan AD, Duoss EB, Stolaroff JK, Giera B. Automated detection and sorting of microencapsulation: Via machine learning. Lab Chip [Internet]. 2019;19(10):1808–17. Available from: 10.1039/C8LC01394B [DOI] [PubMed] [Google Scholar]
- 24.Heo YJ, Lee D, Kang J, Lee K, Chung WK. Real-time Image Processing for Microscopy-based Label-free Imaging Flow Cytometry in a Microfluidic Chip. Sci Rep [Internet]. 2017;7(1):11651. Available from: 10.1038/s41598-017-11534-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitta N, Sugimura T, Isozaki A, Mikami H, Hiraki K, Sakuma S, Iino T, Arai F, Endo T, Fujiwaki Y, Fukuzawa H, Hase M, Hayakawa T, Hiramatsu K, Hoshino Y, Inaba M, Ito T, Karakawa H, Kasai Y, Koizumi K, Lee SW, Lei C, Li M, Maeno T, Matsusaka S, Murakami D, Nakagawa A, Oguchi Y, Oikawa M, Ota T, Shiba K, Shintaku H, Shirasaki Y, Suga K, Suzuki Y, Suzuki N, Tanaka Y, Tezuka H, Toyokawa C, Yalikun Y, Yamada M, Yamagishi M, Yamano T, Yasumoto A, Yatomi Y, Yazawa M, Carlo D Di, Hosokawa Y, Uemura S, Ozeki Y, Goda K, Di Carlo D, Hosokawa Y, Uemura S, Ozeki Y, Goda K. Intelligent Image-Activated Cell Sorting. Cell [Internet]. 2018;175(1):266–276.e13. Available from: http://www.sciencedirect.com/science/article/pii/S0092867418310444 [DOI] [PubMed] [Google Scholar]
- 26.Gupta A, Harrison PJ, Wieslander H, Pielawski N, Kartasalo K, Partel G, Solorzano L, Suveer A, Klemm AH, Spjuth O, Sintorn I-MM, Wählby C. Deep Learning in Image Cytometry: A Review. Cytom Part A [Internet]. 2019;95(4):366–80. Available from: 10.1002/cyto.a.23701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Ouyang M, Ray A, Liu T, Kong J, Bai B, Kim DD, Guziak A, Luo Y, Feizi A, Tsai K, Duan Z, Liu X, Kim DD, Cheung C, Yalcin S, Ceylan Koydemir H, Garner OB, Di Carlo D, Ozcan A. Computational cytometer based on magnetically modulated coherent imaging and deep learning. Light-Sci Appl [Internet]. 2019;8(1):91. Available from: 10.1038/s41377-019-0203-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Mahjoubfar A, Chen CL, Niazi KR, Pei L, Jalali B. Deep Cytometry: Deep learning with Real-time Inference in Cell Sorting and Flow Cytometry. Sci Rep [Internet]. 2019;9(1):11088. Available from: 10.1038/s41598-019-47193-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ignatov A Real-time human activity recognition from accelerometer data using Convolutional Neural Networks. Appl Soft Comput [Internet]. 2018;62:915–22. Available from: http://www.sciencedirect.com/science/article/pii/S1568494617305665 [Google Scholar]
- 30.Bresch E, Großekathöfer U, Garcia-Molina G. Recurrent Deep Neural Networks for Real-Time Sleep Stage Classification From Single Channel EEG. Front Comput Neurosci [Internet]. 2018;12:85. Available from: 10.3389/fncom.2018.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiranyaz S, Ince T, Gabbouj M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans Biomed Eng. 2016;63(3):664–75. [DOI] [PubMed] [Google Scholar]
- 32.Lekha S, S. M, Suchetha M Real-Time Non-Invasive Detection and Classification of Diabetes Using Modified Convolution Neural Network. IEEE J Biomed Health Inform. 2018;22(5):1630–6. [DOI] [PubMed] [Google Scholar]
- 33.Wang N, Liu R, Asmare N, Chu C-HH, Sarioglu AF. Processing code-multiplexed Coulter signals via deep convolutional neural networks. Lab Chip [Internet]. 2019;19(19):3292–304. Available from: 10.1039/C9LC00597H [DOI] [PubMed] [Google Scholar]
- 34.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature [Internet]. 1986;323(6088):533–6. Available from: 10.1038/323533a0 [DOI] [Google Scholar]
- 35.Hopfield JJ. Neural networks and physical systems with emergent collective computational abilities. Proc Natl Acad Sci U S A [Internet]. 1982;79(8):2554–8. Available from: https://www.pnas.org/content/79/8/2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demierre N, Braschler T, Linderholm P, Seger U, van Lintel H, Renaud P. Characterization and optimization of liquid electrodes for lateral dielectrophoresis. Lab Chip. 2007;7(3):355–65. [DOI] [PubMed] [Google Scholar]
- 37.Caselli F, Ninno A De, Reale R, Businaro L, Bisegna P, De Ninno A, Reale R, Businaro L, Bisegna P, Ninno A De, Reale R, Businaro L, Bisegna P. A novel wiring scheme for standard chips enabling high-accuracy impedance cytometry. Sens Actuator B-Chem [Internet]. 2018;256:580–9. Available from: 10.1016/j.snb.2017.10.113 [DOI] [Google Scholar]
- 38.Caselli F, Reale R, Nodargi NA, Bisegna P. Numerical Investigation of a Novel Wiring Scheme Enabling Simple and Accurate Impedance Cytometry. Micromachines. 2017;8(9):283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hochreiter S, Schmidhuber J. Long Short-Term Memory. Neural Comput [Internet]. 1997;9(8):1735–80. Available from: 10.1162/neco.1997.9.8.1735 [DOI] [PubMed] [Google Scholar]
- 40.Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw [Internet]. 2005;18(5):602–10. Available from: http://www.sciencedirect.com/science/article/pii/S0893608005001206 [DOI] [PubMed] [Google Scholar]
- 41.Caselli F, Bisegna P. Simulation and performance analysis of a novel high-accuracy sheathless microfluidic impedance cytometer with coplanar electrode layout. Med Eng Phys. 2017;48:81–9. [DOI] [PubMed] [Google Scholar]
- 42.Shen Y, Yalikun Y, Tanaka Y. Recent advances in microfluidic cell sorting systems. Sens Actuator B-Chem [Internet]. 2019;282:268–81. Available from: http://www.sciencedirect.com/science/article/pii/S0925400518319798 [Google Scholar]
- 43.Rashid KM, Louis J. Times-series data augmentation and deep learning for construction equipment activity recognition. Adv Eng Inform [Internet]. 2019;42:100944. Available from: http://www.sciencedirect.com/science/article/pii/S1474034619300886 [Google Scholar]
- 44.Reale R, De Ninno A, Businaro L, Bisegna P, Caselli F. Electrical measurement of cross-sectional position of particles flowing through a microchannel. Microfluid Nanofluid. 2018;22(4):1–13. [Google Scholar]
- 45.De Ninno A, Errico V, Bertani FR, Businaro L, Bisegna P, Caselli F. Coplanar electrode microfluidic chip enabling accurate sheathless impedance cytometry. Lab Chip. 2017;17(6):1158–66. [DOI] [PubMed] [Google Scholar]
- 46.Haandbaek N, Burgel SC, Heer F, Hierlemann A. Characterization of subcellular morphology of single yeast cells using high frequency microfluidic impedance cytometer. Lab Chip. 2014;14(2):369–77. [DOI] [PubMed] [Google Scholar]
- 47.De Ninno A, Reale R, Giovinazzo A, Bertani FR, Businaro L, Bisegna P, Matteucci C, Caselli F. High-throughput label-free characterization of viable, necrotic and apoptotic human lymphoma cells in a coplanar-electrode microfluidic impedance chip. Biosens Bioelectron [Internet]. 2019;111887. Available from: http://www.sciencedirect.com/science/article/pii/S0956566319309662 [DOI] [PubMed] [Google Scholar]
- 48.Rollo E, Tenaglia E, Genolet R, Bianchi E, Harari A, Coukos G, Guiducci C. Label-free identification of activated T-lymphocytes through tridimensional microsensors on chip. Biosens Bioelectron. 2017;94:193–9. [DOI] [PubMed] [Google Scholar]
- 49.Shaker M, Colella L, Caselli F, Bisegna P, Renaud P. An impedance-based flow micro-cytometer for single cell morphology discrimination. Lab Chip. 2014;14(14):2548–55. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Z, Frey O, Franke F, Haandbæk N, Hierlemann A. Real-time monitoring of immobilized single yeast cells through multifrequency electrical impedance spectroscopy. Anal Bioanal Chem [Internet]. 2014;406(27):7015–25. Available from: 10.1007/s00216-014-7955-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu BY, Elbuken C, Shen C, Huissoon JP, Ren CL. An integrated microfluidic device for the sorting of yeast cells using image processing. Sci Rep [Internet]. 2018;8(1):3550. Available from: 10.1038/s41598-018-21833-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honrado C, Ciuffreda L, Spencer D, Ranford-Cartwright L, Morgan H. Dielectric characterization of Plasmodium falciparum-infected red blood cells using microfluidic impedance cytometry. J R Soc Interface [Internet]. 2018. October 31;15(147):20180416. Available from: 10.1098/rsif.2018.0416 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


