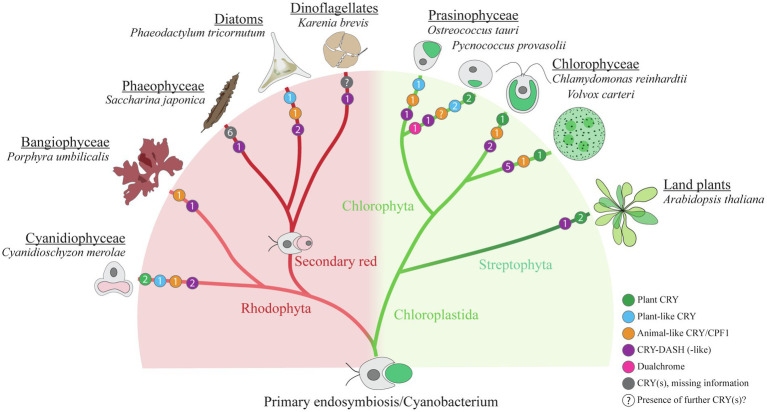
Figure 3.
Cryptochrome distribution within selected algal species. Simplified schematic cladogram of cryptochrome (CRY) distribution within selected algal species in comparison with the land plant Arabidopsis thaliana. Colored circles indicate the numbers (presented in each circle) and representatives of different types of CRYs within the presented species (green, plant CRY; blue, plant-like CRY; orange, animal-like CRY/CPF1; purple, CRY-DASH and CRY-DASH-like; magenta, dualchrome; gray, CRY(s) that are currently not assigned to the former groups; “?,” indicating that there may be further potential CRY(s) in this organism that have not been identified so far). Information for the presented data was taken from the following references: Cyanidioschyzon merolae (Cm; Brawley et al., 2017), Porphyra umbilicalis (Pum; Brawley et al., 2017), Saccharina japonica (Sj; Deng et al., 2012; Yang et al., 2020), Phaeodactylum tricornutum (Pt; Juhas et al., 2014; Oliveri et al., 2014; Fortunato et al., 2015), Karenia brevis (Kb; Brunelle et al., 2007), Ostreococcus tauri (Ot; Heijde et al., 2010; Kottke et al., 2017), Pycnococcus provasolii (Pp; Makita et al., 2021), Chlamydomonas reinhardtii (Cr; Beel et al., 2012; Kottke et al., 2017), Volvox carteri (Vc; Kottke et al., 2017) and Arabidopsis thaliana (At; Chaves et al., 2011). Please note that three putative CRY-DASHs are predicted for Cm in the analysis of Asimgil and Kavakli (2012). For the genus Karenia, three putative CRY/photolyase members are predicted (Shikata et al., 2019). The number of cryptochrome/photolyase members from an additional dinoflagellate and from three stramenopile radiophytes, which all form noxious red tides (Shikata et al., 2019), has been omitted from the figure for clarity. As there is no data available of CRYs from the secondary green line, this line was omitted. The cladogram was modified after Handrich et al. (2017).