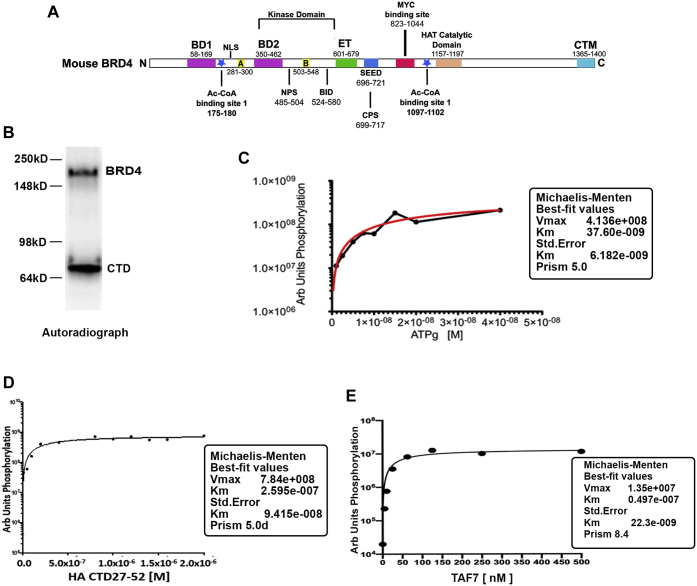
Figure 1.
BRD4 kinase follows classical enzyme kinetics.A, map of mouse BRD4 showing the locations of various domains, the numbers indicate the amino acid residues encompassing the domain. B, in vitro kinase assay using 10uCi radiolabeled ATP showing the auto and trans kinase activities of 10 nM BRD4; 25 nM Pol II CTD27–52 was used as a substrate. Orange line represents the standard error of the best-fit parameters, as determined by Prism. C, Michaelis–Menten plot representing the kinetics of BRD4 autophosphorylation in the presence of increasing concentrations of radiolabeled ATP. D, Michaelis–Menten plot representing the kinetics of 10 nM BRD4 phosphorylation of increasing concentrations of substrate HA-CTD27–52. E, Michaelis–Menten plot representing the kinetics of 10 nM BRD4 phosphorylation of increasing concentration of TAF7 C-terminal fragment. Arbitrary quantification units on Y-axis were plotted against increasing concentrations of TAF7 on X-axis. All Km and Vmax values were calculated as described in Experimental procedures. A and B, conserved motifs; BD1 and BD2, bromodomains 1 and 2; BID, basic residue-rich interaction domain; CTM, C-terminal motif; ET, extra terminal domain; Km, Michaelis–Menten constant; NPS and CPS, N and C-terminal phosphorylation sites respectively; SEED, Ser/Glu/Asp-rich region; Vmax, maximal rate.