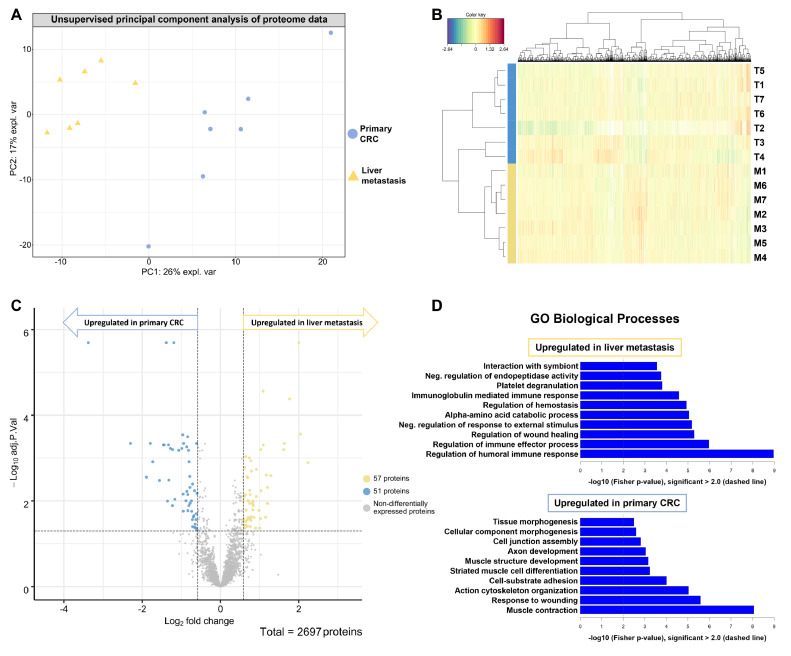
Figure 3.
Unsupervised clustering and statistical analysis reveals distinct proteome biology in primary CRC and corresponding liver metastasis.
The average protein intensity of the two replicates per patient for the primary colorectal carcinoma (T, blue) and the liver metastasis (M, yellow) was computed. Proteins that were at least qualified in 4 out of the 14 samples were used for unsupervised principal component analysis (A) and hierarchical clustering (B). (C) Volcano plot showing proteins with their respective -log10 adjusted p-value and the log2 fold change for the comparison of liver metastases against primary CRC tissue from seven patients. Of the 2697 proteins, 57 were significantly more abundant in liver metastasis, whereas 51 proteins were significantly more abundant in primary colorectal tumors (adjusted p-value < 0.05). (D) Gene ontology (GO) analysis of the significantly dysregulated proteins shows upregulated biological processes for each tumor tissue.