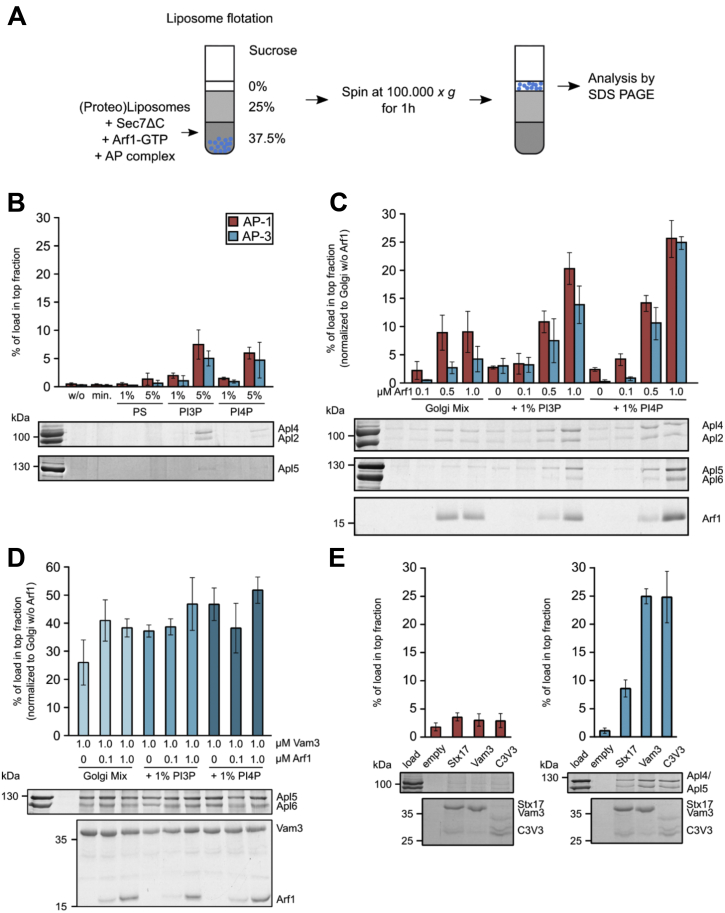
Figure 6.
Cargo, Arf1, and PI4P are required for recruitment of AP-3 to membranes. A, scheme for AP complex flotation assay. Indicated components were incubated with proteoliposomes in the presence of GTP and added to the bottom fraction of a sucrose gradient. Samples were centrifuged and the top fraction was removed for further analyses (see Experimental procedures for details). B, lipids alone have a minor influence on AP-1 and AP-3 recruitment. 0.5 mM of empty liposomes prepared from a minimal lipid mix (73 % DOPC, 18 % DOPE, 8 % Ergosterol, 1 % DAG) with indicated amounts of acidic lipids were incubated with purified AP-1 and AP-3 complex and floated on a sucrose step gradient. The top fraction was removed, proteins were precipitated by TCA, boiled in SDS sample buffer, and analyzed by SDS PAGE. The first lane indicates a sample without liposomes. Band intensity corresponding to Apl4 or Apl5 was determined using FIJI (79). Graphs show the mean percentage of the load in the top fractions. Error bars indicate standard deviation (n = 3). C, Arf1 and PIPs result in a coincidence detection of AP complexes. 0.5 mM of empty liposomes prepared from a Golgi-mimicking lipid mix (see Experimental procedures) and indicated amounts of PIPs were incubated with indicated amounts of Arf1 and purified AP-1 and AP-3 complex and floated on a sucrose step gradient. The top fraction was removed, proteins were precipitated by TCA, boiled in SDS sample buffer, and analyzed by SDS PAGE. Band intensity corresponding to Apl4 or Apl5 was determined using FIJI (79). Graphs show the mean percentage of the load in the top fractions, normalized to a sample using Golgi-Mix liposomes without Arf1 (first band on the gel). Error bars indicate standard deviation (n = 3). D, AP-3 is recruited by Vam3. Empty liposomes or proteoliposomes carrying a total of 1 μM Vam3 were incubated with or without indicated concentrations of Arf1 and purified AP-3 complex and floated on a sucrose step gradient. The top fraction was removed, proteins were precipitated by TCA, boiled in SDS sample buffer, and analyzed by SDS PAGE. Band intensity corresponding to Apl5 was determined using FIJI (79). Graphs show the mean percentage of the load in the top fractions, normalized to a sample using Golgi-Mix liposomes without Arf1 or Vam3 (first band on the gel). Error bars indicate standard deviation (n = 3). E, AP-3 recruitment by cargo is specific. Empty liposomes or proteoliposomes carrying a total of 1 μM Stx17, Vam3 or GST-Chs3(aa1-38)-Vam3(TMD) (C3V3) were incubated with purified AP-1 or AP-3 complex and floated on a sucrose step gradient. The top fraction was removed, proteins were precipitated by TCA, boiled in SDS sample buffer, and analyzed by SDS PAGE. Band intensity corresponding to Apl4 or Apl5 was determined using FIJI (79). Graphs show the mean percentage of the load in the top fractions. Error bars indicate standard deviation (n = 3).