Figure EV5. Design and characterization of protein fragment complementation assay using a split DHFR system.
-
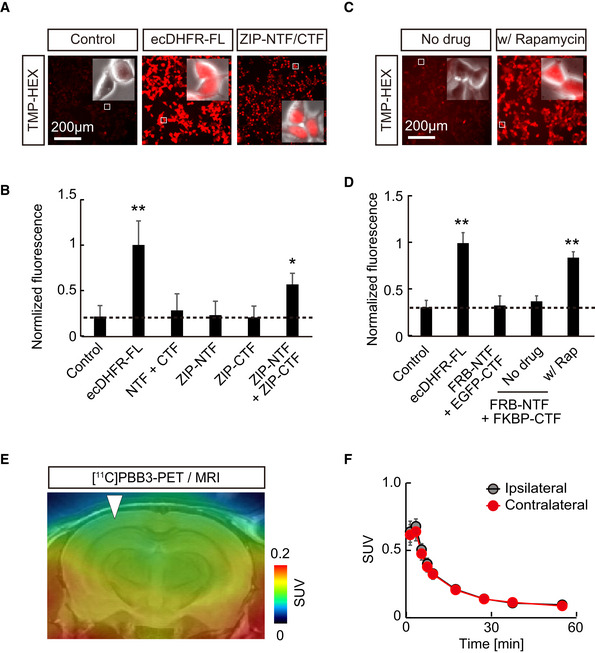
A–DCultured HEK293 cells expressing various constructs were incubated with 100 nM TMP‐HEX and analyzed by fluorescent microscopy. TMP‐HEX efficiently labeled cultured cells co‐expressing NTF and CTF conjugated to a self‐assembling leucine zipper (ZIP) motif, or a rapamycin‐dependent heterodimerization motifs FKBP12‐rapamycin binding domain (FRB) and FK506 binding protein (FKBP) in the rapamycin‐dependent manner, indicating that the cDHFR‐PCA system functions under these conditions as designed. (A) Representative images illustrate a selective retention of TMP‐HEX in cells carrying full‐length ecDHFR (as positive control) or a combination of ZIP‐tagged ecDHFR‐NTF and ecDHFR‐CTF. Insets demonstrate high‐magnification fluorescence images overlaid with phase‐contrast pictures of individual cells. Note that ZIP‐tagged ecDHFR‐NTF and CTF preferentially localize in nucleus. (B) Normalized fluorescence intensities of TMP‐HEX in cells transfected with indicated constructs. Mean value of full‐length ecDHFR (ecDHFR‐FL) was set as 1. Note that only ecDHFR‐FL and the split protein that reassembles via ZIP interaction retain the marker above background levels. Data from six independent experiments are plotted as mean ± SD. F(5, 28) = 21.12; *P < 0.05, **P < 0.01 (one‐way ANOVA followed by Dunnett post hoc test). (C) HEK293 cells expressing various constructs were incubated with 100 nM TMP‐HEX with or without 500 nM rapamycin (LC Laboratories) and imaged by fluorescence microscopy. Representative images illustrate selective labeling of TMP‐HEX in cells co‐expressing ecDHFR‐NTF and CTF tagged with FRB and FKBP in the presence of rapamycin. Insets demonstrate high‐magnification fluorescence photomicrographs overlaid with phase‐contrast images. (D) Rapamycin (Rap)‐induced FRB‐FKBP interaction was assessed as TMP‐HEX labeling efficiency in cells expressing various combinations of ecDHFR‐NTF and ecDHFR‐CTF fragments. Fluorescence intensities were normalized by mean value of labeling of full‐length ecDHFR (ecDHFR‐FL) with TMP‐HEX. Data from four independent experiments are presented as mean ± SD. F(4, 15) = 55.08; **P < 0.01 (one‐way ANOVA followed by Dunnett post hoc test).
-
EA representative coronal PET image captured with [11C]PBB3, a potent PET tracer for aggregated tau fibrils, in mice expressing TRD‐NTF and TRD‐CTF by AAV injection into one side of somatosensory cortex. Averaged image of dynamic scan data at 0‐60 min after i.v. injection of [11C]PBB3 is shown. A template MRI image was overlaid for spatial alignment of the PET image. Arrowhead indicates injection site.
-
FKinetics of [11C]PBB3 in mouse brain during 60‐min dynamic PET scan. VOIs were manually placed on ipsilateral and contralateral cortical areas for quantification. Data from four mice expressing TRD‐NTF and TRD‐CTF are plotted as mean ± SEM. Note there is no significant difference in radioactive signals of [11C]PBB3 between ipsilateral and contralateral sides.
Source data are available online for this figure.
