Figure EV1. The CRL4‐DCAF1 oligomerization state changes upon neddylation and is dependent on LisH, the DCAF1 dimerization domain.
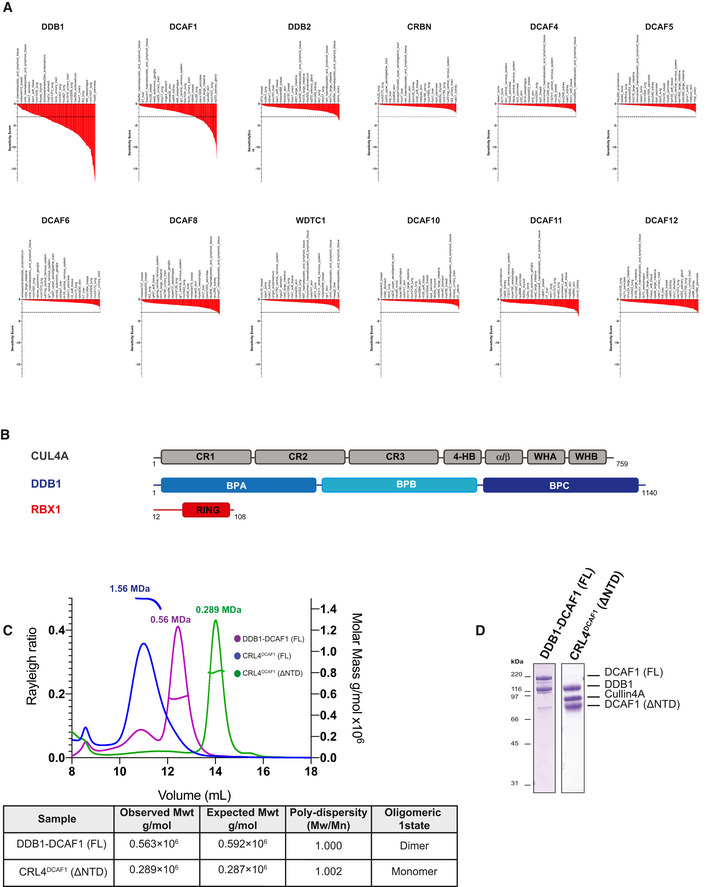
- Sensitivity score profiles adapted from the DRIVE data for DDB1, DCAF1, DDB2, CRBN, DCAF4, DCAF5, DCAF6, DCAF8, WDTC1, DCAF10, DCAF11, and DCAF12. The dropout score in the shRNA screen was converted into log fold‐change (logFC) per shRNA per cell line. The logFC was then normalized per sample to obtain a shRNA level sensitivity score. The shRNA level scores are further aggregated to gene level sensitivity scores using either the ATARiS algorithm (Shao et al, 2013) or the RSA algorithm (König et al, 2007). These give a measure of the statistical significance of the dropout of those 20 shRNAs used per gene compared to the remainder shRNAs in the screen (McDonald et al, 2017).
- Domain organization of human CUL4A, DDB1, and RBX1. CR, cullin repeat; UFD, ubiquitin‐fold domain; RING, really interesting new gene; WH, winged helix; BP, β‐propeller.
- SEC‐MALS analysis, the chromatogram shows the Rayleigh ratio curves of CRL4DCAF1 (FL), CRL4DCAF1(ΔNTD), and DDB1‐DCAF1 (FL) together with the molar mass (MDa) of the main peaks determined by MALS. The table summarizes the SEC‐MALS observed molecular weights, the calculated molecular weight, polydispersity values, and oligomeric states of the tested complexes.
- SDS–PAGE of the corresponding complexes.
