Figure 3. The DCAF1 WD40 domain is essential for tetramer formation.
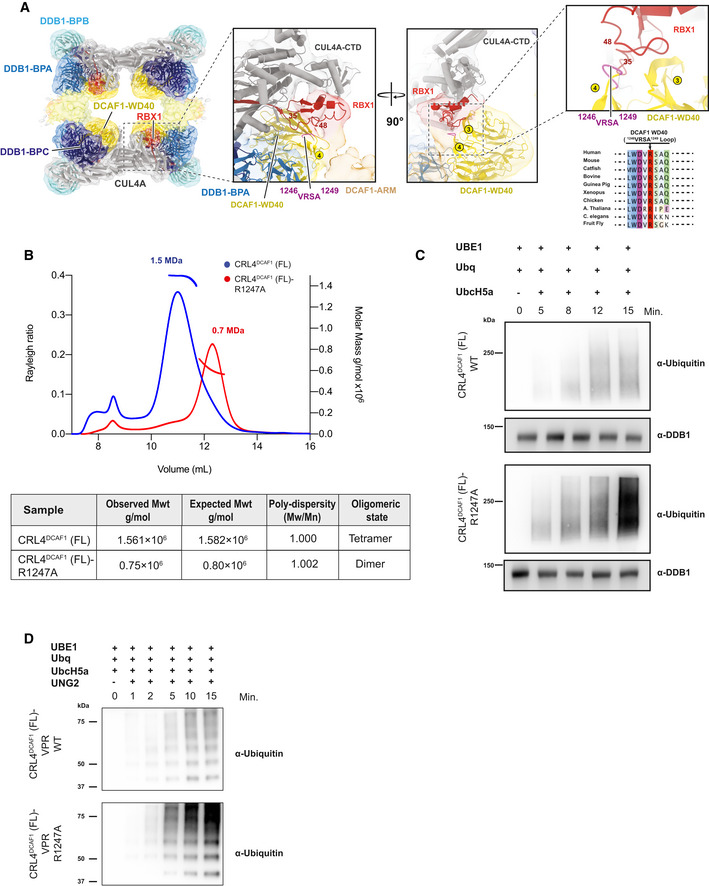
- CRL4DCAF1 cryo‐EM map (8.4 Å) with fitted model (left), and a close‐up view of the CRL4DCAF1 map shows the interaction between DCAF1 (WD40) in yellow and RBX1 in red. The loop in DCAF1 (WD40) that contains the mutated arginine (R1247) is colored in violet. Circled numbers indicate the respective blades in the WD40 β‐propeller. The RBX1 segment potentially contacting the VRSA loop in DCAF1 is indicated by flanking residue numbers. Conservation plot in the region around the residue R1247 in DCAF1 is shown to the lower right.
- SEC‐MALS analysis of CRL4DCAF1 and CRL4DCAF1 (R1247A) mutant. The chromatogram displays Rayleigh ratio curves of CRL4DCAF1 WT (blue) and mutant CRL4DCAF1 (R1247A) (red) together with the molar mass (MDa) of the main peaks calculated by MALS. The table summarizes the SEC‐MALS observed molecular weights in the main peaks, the calculated molecular weight, polydispersity values, and oligomeric states of the tested complexes.
- Autoubiquitination of DCAF1 by wild‐type CRL4DCAF1 (FL) (top) or mutant CRL4DCAF1 (FL) (R1247A) (bottom) observed after incubation with UBA1, UbcH5a, Ubiquitin (WT), at 30°C for 0 to 15 min as indicated (n = 3).
- Ubiquitination of UNG2 by wild‐type CRL4DCAF1‐VPR (FL) (top) or mutant CRL4DCAF1‐VPR (FL) (R1247A) (bottom) observed after incubation with UBA1, UbcH5a, Ubiquitin (WT), at 30°C for 0 to 15 min as indicated (n = 3).
Source data are available online for this figure.
