Abstract
Gliomas are the most common primary intrinsic brain tumors occurring in adults. Of all malignant gliomas, glioblastoma (GBM) is considered the deadliest tumor type due to diffuse brain invasion, immune evasion, cellular, and molecular heterogeneity, and resistance to treatments resulting in high rates of recurrence. An extensive understanding of the genomic and microenvironmental landscape of gliomas gathered over the past decade has renewed interest in pursuing novel therapeutics, including immune checkpoint inhibitors, glioma-associated macrophage/microglia (GAMs) modulators, and others. In light of this, predictive animal models that closely recreate the conditions and findings found in human gliomas will serve an increasingly important role in identifying new, effective therapeutic strategies. Although numerous syngeneic, xenograft, and transgenic rodent models have been developed, few include the full complement of pathobiological features found in human tumors, and therefore few accurately predict bench-to-bedside success. This review provides an update on how genetically engineered rodent models based on the replication-competent avian-like sarcoma (RCAS) virus/tumor virus receptor-A (tv-a) system have been used to recapitulate key elements of human gliomas in an immunologically intact host microenvironment and highlights new approaches using this model system as a predictive tool for advancing translational glioma research.
Keywords: animal modeling, genetically engineered, glioblastoma, high-grade glioma, immunocompetent, patient-derived xenograft, preclinical testing, RCAS/tv-a, tumor microenvironment
1 |. INTRODUCTION
Gliomas are the most common type of primary intrinsic brain tumors occurring in adults. With an annual incidence rate of 7.08 cases per 100,000; malignant brain tumors are the fifth most common cause of cancer-related deaths among adolescents and young adults in the United States (Ostrom et al., 2020). Infiltrating gliomas, the most common subtype of glioma in adults, can present as malignant tumors or evolve to malignancy from lower grade infiltrating astrocytomas and oligodendrogliomas. Even anaplastic forms of well-circumscribed gliomas, such as ependymomas, can show aggressive and malignant growth patterns. Grade IV astrocytoma, also known as glioblastoma (GBM), is a particularly deadly form of glioma (median survival ~15 months) and recurs despite combination surgical, chemotherapy, and radiation treatments (Lapointe, Perry, & Butowski, 2018; Louis et al., 2016; Wesseling & Capper, 2018).
Over the past decade, significant progress has been made in understanding glioma biology. From a histopathological perspective, GBM is characterized by cellular and microvascular proliferation, atypical nuclei, pseudopalisading necrosis, and diffuse infiltration of the surrounding brain parenchyma. The considerable molecular and cellular heterogeneity present in GBMs also licenses them for frequent recurrence due to the high probability for treatment-resistant elements (Omuro & DeAngelis, 2013; Wen & Kesari, 2008). Genomic and transcriptomic profiling of human GBMs have revealed that alterations in three core signaling pathways (receptor tyrosine kinase [RTK]/Ras/PI3K cascades, tumor suppressor p53, and cell cycle regulator Rb) are the major drivers of these tumors, providing a framework for molecular categorization (Brennan et al., 2013; Cloughesy, Cavenee, & Mischel, 2014; Verhaak et al., 2010; Wang et al., 2017). Single-cell transcriptomic studies, however, show that multiple transcriptional subtypes can co-exist in one tumor, possibly due to environmental factors (Neftel et al., 2019; Wang et al., 2017). Studies of the tumor microenvironment (TME) in GBM have revealed their highly immune-suppressive nature owing to high numbers of alternatively activated macrophages, low T-cell populations, and elevated levels of immunosuppressive cytokines. The interactions of tumor cells with neural cell types such as astrocytes, microglia and neurons, and the unique extracellular matrix components, adds another layer of complexity to the microenvironment of these tumors (Quail & Joyce, 2017; Wang et al., 2017).
Animal models that can replicate the molecular, histopathological, and immunological features of the human disease are a central requirement for predictive preclinical testing. Many new therapies have shown promise during preliminary stages of research but failed in clinical trials in part due to differences in the biological properties of animal models and the human TME (Lenting, Verhaak, Ter Laan, Wesseling, & Leenders, 2017). Currently, various orthotopic, syngeneic as well as patient-derived xenograft (PDX) models of glioma are used for preclinical testing, but these models each have their own limitations and do not always faithfully reflect the characteristics of human GBM (Chen, Zhang, Yang, Hagan, & Li, 2013; Huszthy et al., 2012; Stylli, Luwor, Ware, Tan, & Kaye, 2015). There are several key features that should be present as genetic components of a clinically relevant animal model of human glioma: (a) genomic alterations should resemble those that are frequently found in human gliomas, (b) these alterations should be introduced to only a limited number of cells, (c) mutant genes should be expressed within a specific target tissue or cell type, and (d) the genetic alterations should mirror the gene expression hierarchies characteristic of human glioma genes (e.g., embryonic developmental programs) (Lenting et al., 2017). Several reviews on animal models of glioma have described the advantages of using genetically engineered rodent models over other commonly used syngeneic and PDX models (Fomchenko & Holland, 2006; Miyai et al., 2017). Genetically engineered models (GEMs) provide a useful platform to induce different tumorigenic genomic alterations and specify the cell types to undergo transformation, all in an immunologically intact microenvironment, which recapitulates tumor-immune cell interactions found to be important drivers of disease progression (Hambardzumyan, Parada, Holland, & Charest, 2011; Huse & Holland, 2009; Oh et al., 2014).
Numerous strategies have been used to establish GEMs of gliomas in rodents. Here we detail the retrovirus-based RCAS/tv-a system, which enables spatial and temporal control of genetic transformations in the brain to mimic many of the key features of gliomas in immunocompetent hosts. This review summarizes the characteristics of the RCAS/tv-a glioma model system and discusses new and instructive examples that have successfully used this tool for faithful glioma modeling to study tumor pathobiology and test new therapeutic approaches.
2 |. OVERVIEW OF THE RCAS/TV-A SYSTEM
2.1 |. Features of the RCAS vector
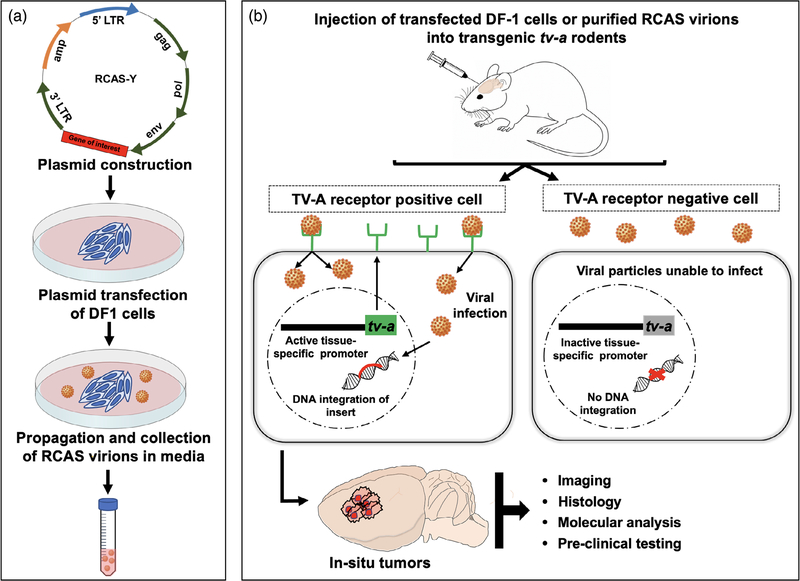
The RCAS vector belongs to the avian leukosis virus-A subgroup of retroviruses. The vector was designed by substituting the src gene of the well-known oncogenic Rous sarcoma virus (RSV) with a multicloning site for inserting gene-coding regions of interest (generally ≤2.5 kb). The 3′ end of the RSV src gene has a splice acceptor site which has been retained in the RCAS vector. This site regulates the expression of the cloned gene of interest under the control of the viral LTR promoter which is how the vector derives the name—RCAS— Replication-Competent ASLV long terminal repeat with Splice acceptor (Orsulic, 2002). RCAS vectors are replication-competent only in avian cells and not in mammalian cells. Hence, an immortalized chicken fibroblast cell line, DF-1, is typically used to propagate and maintain high-titer stocks of the virus (Figure 1a). The viral yield can be further boosted by replacing the pol region with one from the Bryan high-titer strain of RSV. This form of RCAS is referred to as RCASBP and is preferred for in vivo infections (Ahronian & Lewis, 2014; Li et al., 2012) owing to its high titer capacity and stronger insert expression. Although the insert size is limited to ~2.5 kb, a variety of oncogenes (Dai et al., 2001; Holland et al., 2000; Ozawa et al., 2014; Ozawa et al., 2018; Shih et al., 2004; Szulzewsky et al., 2020), genes encoding dominant-negative and truncated forms of proteins (Chen et al., 2020; Du & Li, 2007; Ene et al., 2020; Holland, Hively, DePinho, & Varmus, 1998), miRNAs (Huse et al., 2009) and shRNAs (Bromberg-White et al., 2004; Seidler et al., 2008) have been successfully cloned into RCAS vectors to study their role in tumorigenesis. Insert size limitations can also be overcome by eliminating some of the viral structural genes such as env (e.g., BBAN vectors) (Li et al., 2012) (see Section 3 below). RCAS vectors have also been used to deliver bioluminescence and fluorescence-based reporter gene constructs to enable in vivo imaging and tracking of tumors and specific cell types (Becher et al., 2010; L. Uhrbom, Nerio, & Holland, 2004; Halliday et al., 2014). Higher efficiency, specificity, and control of gene expression have been achieved by adding internal promoters to the vectors (e.g., RCAN vectors) (Li et al., 2012). Such ease of customization makes the RCAS vector adaptable for modeling a wide range of cancers in vivo (Ahronian & Lewis, 2014).
FIGURE 1.
Schematic depiction of the RCAS/tv-a glioma model. (a) DF-1 chicken fibroblasts are transfected with RCAS plasmids carrying a gene of interest for propagation in vitro. (b) Transfected DF-1 cells or the isolated RCAS virions are injected into transgenic tv-a rodent hosts where they selectively infect TV-A-positive cells, in some instances confined by cell -or tissue-specific tv-a gene expression. Stable integration of the gene of interest into host DNA then results in tumors with specific features. Abbreviations: RCAS, replication-competent ASLV long terminal repeat splice acceptor; tv-a, tumor virus receptor A
2.2 |. The TV-A receptor
Another advantage of the RCAS/tv-a system is the spatiotemporal control of tv-a gene expression, which is governed by the presence of the tumor virus A (TV-A) protein receptor on the surface of the target host cells (Figure 1b). The viral envelope glycoproteins bind to the TV-A receptor and enter host cells. Avian cells naturally express TV-A protein on their surface making them susceptible to infection. Mammalian cells, which otherwise lack this receptor, can be engineered to express the tv-a gene under the control of tissue-specific promoters thereby localizing its expression only within the cells where the selected promoter is activated (Li et al., 2012). This additional layer of control provides more scientific flexibility in exploring gene transformations.
The DF-1 cells transduced with RCAS vectors are typically used to deliver viral particles to TV-A positive cells in vitro or in vivo. Since the virus is unable to enter and replicate in mammalian cells, there is a minimum risk of cell-to-cell spread of infection. The TV-A-positive host cells are also susceptible to multiple rounds of infection which enables the use of several RCAS vectors carrying different target genes or shRNAs either simultaneously or sequentially at any time during the experiment (Ahronian & Lewis, 2014; Ozawa et al., 2014). This enables the evaluation of multiple gene combinations on cancer development using a single tv-a transgenic rodent line rather than generating and maintaining separate and/or cross transgenic lines for genes of interest. Using tetracycline-induction, cre-lox recombinases, or selecting temporally activated promoters offers additional control over the desired gene expression (Ahronian & Lewis, 2014; Hambardzumyan, Amankulor, Helmy, Becher, & Holland, 2009; Mayr et al., 2008). As cancer is frequently a disease of multiple genetic alterations, initially developing in relatively few “cells of origin” and then acting in synergy to drive tumorigenesis, these properties of the RCAS/tv-a system have made it an important means of accurately modeling cancer progression.
2.3 |. tv-a transgenic animals
To adapt the RCAS/tv-a technique for in vivo tumor modeling, transgenic mice expressing the TV-A receptor under various tissue-specific promoters have been generated. Some examples include keratin-tv-a, elastase-tv-a, or stem cell leukemia (scl)—tv-a (SCL-tv-a) transgenic mice that have been used for in vivo modeling of breast and ovarian cancer, pancreatic cancer, and hemangiomas, respectively (Fisher et al., 1999; Li et al., 2012; Von Werder, Seidler, Schmid, Schneider, & Saur, 2012). Holland et al. were the first to generate two strains of transgenic mice for specifically modeling gliomas; one with glial fibrillary acidic protein (GFAP) promoter-driven tv-a (Gtv-a) expression targeting both immature and mature astrocytes and the other with nestin promoter-driven tv-a (Ntv-a) expression targeting neural progenitor cells (Holland et al., 1998; Holland & Varmus, 1998). These tv-a transgenic mice have also been crossed with mice bearing germline mutations in tumor suppressors (e.g., p53, PTEN) or cell cycle arrest proteins (e.g., INK4a-Arf/CDKN2A, Rb) commonly implicated in glioma progression, to study their combined effect on tumorigenesis (Brennan et al., 2013; Li et al., 2012).
Breeding with luciferase-expressing transgenic reporter mice has enabled in vivo monitoring of tumor cells via bioluminescence imaging (BLI) (Uhrbom et al., 2004; Becher et al., 2008). For example, Uhrbom and colleagues crossbred the Ntv-a mice with Ef-luc reporter mice, which expressed luciferase under the tumor-specific human E2f1 promoter, to generate Ef-luc-Ntv-a double transgenic mice (Uhrbom et al., 2004). Following injection of RCAS-PDGF-B viruses, BLI was used to assess tumor growth in the mice and to monitor response to treatment by PDGFR and mTOR inhibitors. A similar approach was adopted by Becher et al. in a study examining the role of sonic hedgehog (SHH) signaling in PDGF-B-induced gliomas (Becher et al., 2008). They generated Gli-luc-Ntv-a mice, where luciferase was expressed under a glioma-associated oncogene homolog (Gli)-responsive promoter. Luciferase expression could also be triggered by the injection of RCAS-Cre in tv-a transgenic mice bearing the Lox-Stop-Lox luciferase cassette (Ene et al., 2020).
In addition to luciferase, many fluorescence reporter mice (GFAP-GFP+, Olig2+-GFP+, Cx3cr1GFP/wt-Ccr2RFP/wt) have been crossed with various tv-a transgenic mice for in vivo tracing, downstream cell isolation, and functional analyses of specific cell types in the glioma microenvironment (Alexander et al., 2020; Halliday et al., 2014; Szulzewsky et al., 2015).
Newer tv-a strains driven by promoters active in specific cell populations in the brain during various stages of neurodevelopment (e.g., Pax3-tv-a, Ctv-a, Btv-a) have aided the study of the origin and biology of different CNS tumors such as pediatric gliomas and ependymomas (Lindberg, Kastemar, Olofsson, Smits, & Uhrbom, 2009; Misuraca, Hu, Barton, Chung, & Becher, 2016; Ozawa et al., 2018). More recently, our group has generated Ntv-a transgenic rats, further extending the application of this brain tumor modeling technology to an additional rodent species (Connolly et al., 2017; Connolly et al., 2018). As the understanding of neurobiology and the processes underlying gliomagenesis evolves, novel strains of tv-a rodents could be created for use with the increasing variety of RCAS vectors.
3 |. ADVANTAGES AND DISADVANTAGES OF THE RCAS/tv-a SYSTEM
Accurately modeling gliomas in vivo comes with distinct requirements that are met efficiently using the RCAS/tv-a system (Table 1). For example, gliomas form in situ without the intracranial injection of tumor cells. Importantly, dividing and non-dividing cells can be infected in the brain and, uniquely, neural precursor cells can specifically be targeted for studying gliomagenesis. Such a controlled pattern of infection faithfully models the clonal origin of solid tumors and a core, driving population of cancer stem-like cells. More generally, the RCAS/tv-a system allows for tumor modeling in immunocompetent hosts. The RCAS/tv-a system, paired with CRISPR-Cas9 technologies, enables the introduction of loss-of-function point mutations or truncations into the sequence of the gene of interest encoded in the RCAS plasmid, to directly map the functional importance of protein domains for the oncogenic capabilities of said genes (Ozawa et al., 2018; Szulzewsky et al., 2020).
TABLE 1.
Advantages and disadvantages of commonly used animal models of glioma
| Model | Advantages | Disadvantages |
|---|---|---|
| Patient-derived xenograft (PDX) | Mimic heterogeneity observed in human tumors | Varying degree of immunodeficiency in hosts limits study of tumor-immune microenvironment interactions |
| Humanized hosts can offer a more realistic microenvironment | Serial passaging over time can result in loss of original tumor characteristics | |
| High penetrance rates and shorter latency periods | Tumors do not form in situ, hence not suitable for studying early events in tumorigenesis | |
| Minimum animal breeding and colony maintenance requirements | Lack of spatial or temporal control on tumorigenesis | |
| Spontaneous and/or inducible GEMs (non-RCAS) | Tumors form in situ (spontaneous GEMs), hence useful for understanding events occurring in early tumorigenesis | Variable penetration and tumor latencies |
| Inducible GEMs (e.g., Cre-inducible) allow spatiotemporal control on tumorigenesis | Requires extensive animal breeding and genotype monitoring protocols | |
| Immunocompetent hosts provide intact microenvironment | Less specificity of targeting cells/tissues of interest | |
| Modifiable RCAS/tv-a GEMs | High spatial and temporal control with local delivery of virus and promoter-driven tv-a expression | Limited gene insert size capacity of the virus (∼2.5 kb) |
| Regional and dose-controlled infectivity mimic clonal origin of human solid tumors | Variable infectivity can also limit the investigation to highly penetrant oncogenes | |
| Versatile tumor modeling using a single tv-a rodent strain | Requires animal breeding and genotype monitoring protocols | |
| Efficient infection of non-dividing cells minimizes off-target effects | The higher cost of animal colony maintenance | |
| Robust for in vitro and in vivo infection/modeling | Variable penetration and tumor latencies in some contexts | |
| Powerful synergies with existing gene-editing tools allow for engineering complex gene fusions, translocations, etc. (e.g., Cre-loxP, CRISPR-Cas9) | ||
| Immunocompetent hosts |
Abbreviations: GEM, genetically engineered model; RCAS, replication-competent ASLV long terminal repeat with splice acceptor; tv-a, tumor virus a.
A key limitation of the RCAS/tv-a model comes with its relatively small gene insert size capacity. Some efforts have been made to customize the vector by deleting certain viral structural elements (e.g., env-deleted BBAN vectors) to accommodate inserts larger than 2.5 kb, but these experiments have resulted in replication-incompetent vectors which yield significantly lower viral titers, rendering them unfeasible for in vivo infections (Chen et al., 1999; Li et al., 2012). Nevertheless, multiple alternate approaches have been adopted to circumvent this problem. An example of this is the use of RCAS/tv-a models to study the role of constitutively active EGFR mutants (EGFRvIII) in glioma biology. The large size of the EGFR gene does not permit its incorporation into the RCAS plasmid. Hence, a truncated version of the EGFR cDNA that would retain the constitutive kinase activity of the mutant receptor was used to construct an RCAS vector (Holland et al., 1998). More recent studies have used transgenic EGFRvIIIfl-stop-fl mice injected with RCAS vectors carrying cre-recombinases to model EGFRvIII-driven gliomas (Chen et al., 2020; Ene et al., 2020). Some groups have also used modified retroviral or lentiviral vectors pseudotyped with ASLV-A (Env A), the ligand for TV-A receptor, to deliver large genes while still retaining the specificity of infection offered by RCAS/tv-a (Von Werder, Seidler, Schmid, Schneider, & Saur., 2012; Ahronian & Lewis., 2014).
Another limitation of the RCAS/tv-a system is that in some cases the generated tumors can have very low penetrance and long latency periods, depending on the oncogene used. This may limit some investigations to only specific genes or gene combinations which result in higher penetrance rates.
4 |. VERSATILE TUMOR MODELING USING THE RCAS/TV-A SYSTEM
The generation of multiple transgenic tv-a rodent strains has greatly expanded the utility of RCAS/tv-a as a tool for tumor modeling. Furthermore, genetic manipulations such as introducing bioluminescence reporters (Uhrbom et al., 2004, Becher et al., 2008; Ene et al., 2020), fluorescence reporters (Becher et al., 2010; Lindberg et al., 2009; Alexander et al., 2020; Halliday et al., 2014), and cell barcodes (Sanz et al., 2009) have not only enabled in vivo tumor imaging and tracking of individual cell lineages but also expanded the scope of performing a wide variety of genomic and proteomic analyses and measuring response to different treatment modalities. The following sections of this review summarize the various ways in which RCAS/tv-a-driven glioma models have served as a versatile platform for studying glioma biology and preclinical testing of novel therapeutics.
4.1 |. Understanding the molecular drivers of glioma initiation
Due to the high degree of molecular and cellular heterogeneity in human gliomas, establishing faithful in vivo models has been a formidable challenge. The use of the RCAS/tv-a system has been helpful in delineating numerous aspects of glioma biology, especially regarding immunology, given the in-situ tumor formation in immunocompetent hosts. In such early work, it was demonstrated that specific co-operative effects of oncogenic RTK signaling and loss of function of cell cycle arrest pathways promote glioma progression in mice (Holland et al., 1998). For example, in an early model, only mice with combined oncogenic mutations of both RTK and cell cycle pathways (in this study, constitutive EGFR activation and INK4a-Arf deletion, respectively) developed glioma-like lesions resembling human disease. The co-operative effects of constitutive EGFR activity, p53 deletion, and CDK4 overexpression were also shown to generate gliomas in Ntv-a mice (Holland et al., 1998). Using a similar approach, the role of other growth factors, such as fibroblast growth factor (FGF)-2, platelet-derived growth factor (PDGF)-B, and insulin-like growth factor- binding protein (IGFBP)-2, in promoting and maintaining high-grade gliomas was demonstrated (Holland & Varmus, 1998; Holmes et al., 2012; Shih et al., 2004). These studies have also helped elucidate the redundancy and crosstalk derived from different oncogenic mutations and signal transduction pathways (e.g., MAPK, JAK–STAT, NF-κB, integrins) driving glioma progression (Gressot et al., 2015; Holmes et al., 2012; Robinson et al., 2010).
The versatility and efficiency of the RCAS/tv-a system have been demonstrated by validating computational and mathematical analyses of human GBM data in mouse models to connect the three GBM transcriptional molecular subtypes (classical, proneural, and mesenchymal) in an evolutionary framework. Ozawa et al. reported that more primitive tumors were “proneural” in character (Ozawa et al., 2014). In both Ntv-a and Gtv-a mice, overexpression of PDGF-A, predicted by computational analyses to be the strongest initial driving event in glioma evolution, was one of the only single transformations sufficient to drive gliomagenesis in vivo. The combination of PDGF-A and Tp53 loss recreated by injecting RCAS-PDGF-A and RCAS- shp53-RFP was able to induce high-grade gliomas in 100% of injected Ntv-a and Gtv-a transgenic mice. Similarly, RCAS-shp53-RFP and RCAS-shNf1-GFP vectors injected together in Ntv-a and Gtv-a transgenic mice, generated gliomas with genomic and histological features resembling the mesenchymal transcriptional subtype. These models were also used to validate many other conclusions of the human data-based computational and mathematical framework (e.g., shortening of survival in PTEN deficient, PDGF-A-induced gliomas; PDGF-A elevation followed by CDKN2A loss propagates tumor formation; and late loss of NF1 converts PDGF-A-induced proneural tumors to those with a mesenchymal gene expression pattern) (Ozawa et al., 2014). Oldrini et al. have further expanded the utility of this technique by combining more sophisticated gene-editing tools like CRISPR-Cas9 (see below) to recreate more complex gene fusions and chromosomal deletions commonly encountered in certain rare gliomas (Oldrini et al., 2018). Finally, Pattwell et al. showed that a splice variant of NTRK2/TrkB (TrkB.T1) which lacks the intracellular kinase domain of full-length TrkB is specifically upregulated in glioma tumors compared with normal brain. Combined expression of RCAS-TrkB.T1 with RCAS-PDGF-B resulted in a significantly shorter tumor latency compared with the expression of RCAS-PDGF-B alone (Pattwell et al., 2020).
Another significant contribution of the RCAS/tv-a technology has been in the field of pediatric brain tumor research. RCAS vectors were used to develop the first genetically engineered mouse model for diffuse intrinsic pontine glioma (DIPG), a rare and incurable form of pediatric brain tumor originating in the brainstem (Misuraca, Cordero, & Becher, 2015). Limited access to patient tumor samples and a lack of understanding of tumor biology has been a deterrent to the development of any new treatments for DIPG. However, this model, established by delivering RCAS-PDGF-B to the brain stem of Ink4a-Arf −/− neonatal mice, demonstrated the transforming potential of cells outside the subventricular zone in the brain and effectively mimicked key histological features of the human disease (Becher et al., 2010). In subsequent years, additional RCAS-driven models of DIPG integrating the most common genetic and epigenetic mutations of the disease (i.e., PDGF overexpression, p53 loss, and the H3.3K27M mutation) were engineered (Misuraca et al., 2015). A newer model for DIPG using Pax3-tv-a transgenic mice, where Pax3 is a transcription factor specific for cells located in the brainstem, was also established. These mice, injected with RCAS-PDGF-B, RCAS-Cre (for p53 deletion), and/or RCAS-H3.3K27M, developed varying degrees of low-grade and high-grade gliomas with histological characteristics resembling human DIPG tumors (Misuraca et al., 2016).
4.2 |. Exploring the “cells-of-origin” in gliomas
Cells of origin in cancer are defined as cells that either inherently have or have acquired transforming alterations (e.g., inherited or somatic gene mutations) and initiate a particular type of cancer (Visvader, 2011). The cell(s) of origin in gliomas has been a topic of much debate and study. Over the years, as our understanding of development and neurobiology have evolved, many studies have exploited the cell-specific targeting properties of the RCAS/tv-a system to explore the topic of “cells-of-origin” in gliomas. Most of these studies have targeted delivery of RCAS viruses carrying specific oncogenes mainly to two cell types in the mouse brain (astrocytes and neural progenitors using Gtv-a or Ntv-a mice, respectively). Holland et al. found that RCAS-mediated combined delivery of Akt and Ras oncogenes induced high-grade gliomas in Ntv-a mice but not in Gtv-a mice (Holland et al., 2000). In a follow-up study, it was observed that K-ras alone was sufficient to induce tumor formation in Ink4a-Arf null mice of both Ntv-a and Gtv-a strains, highlighting the role of specific combinations of oncogenic alterations that could promote tumor formation by transforming susceptible cell types in the brain (Lene Uhrbom et al., 2002). This was further exemplified when Lindberg et al. successfully demonstrated glioma formation from more differentiated oligodendrocyte progenitor cells (OPCs) via targeted intracranial delivery of RCAS-PDGF-B in Ctv-a mice where the TVA receptor expression is driven by the OPC-specific 2′, 3′-cyclic nucleotide 3′-phosphodiesterase (Cnp) promoter (Lindberg et al., 2009). OPCs were also found to be susceptible to transformation by Kras-Akt in Ctv-a mice with Arf or Ink4a-Arf deletions (Lindberg et al., 2014). Thus, while most literature suggests that neural progenitors may be the tumor-initiating cells of origin in gliomas, the capability of inducing specific oncogenic alterations to transform more differentiated cell types still warrants further investigation into how gliomas originate. Combining the RCAS/tv-a technology with more sophisticated gene editing and lineage tracing tools may provide a means to advance such investigations.
4.3 |. Modeling isocitrate dehydrogenase wild-type and mutant gliomas
A genome-wide analysis of gliomas revealed that the IDH1 gene is the most commonly mutated gene in low-grade gliomas (grade II-III) and secondary GBMs (Agnihotri et al., 2013). Isocitrate dehydrogenase (IDH) is an enzyme in the citric acid cycle that, when mutated, permits the conversion of isocitrate to the oncometabolite 2-Hydrodxyglutarate (2-HG) and inhibits DNA de-methylation. Thus, IDH-mutant gliomas develop a glioma-CpG island methylator phenotype (G-CIMP), a state of epigenetically altered gene expression via DNA hypermethylation (Noushmehr et al., 2010). Patients with IDH-mutant GBM have significantly longer survival (~31 months) than those with IDH-wild-type tumors (~15 months) (Yan et al., 2009). Hence, now along with histological grade, the IDH status of tumors is also used to classify and predict patient prognosis as these mutations are drivers of less aggressive diffuse astrocytomas and oligodendrogliomas (Cancer Genome Atlas Research et al., 2015; Louis et al., 2016; Yan et al., 2009).
Several groups have attempted to develop animal models of IDH-mutant gliomas, which have proved to be a challenging task due to the perinatal lethality of conditional knock-in of mutant IDH1 using nestin-Cre or GFAP-Cre (Pirozzi et al., 2017). Tamoxifen-induced activation of IDH1R132H restricted to the subventricular zone (SVZ) of adult mice does not result in gliomas, suggesting that the IDH mutation alone is not sufficient for gliomagenesis (Philip et al., 2018). Amankulor et al. were the first to develop IDH-wt and IDH-mutant mouse models using the RCAS/tv-a system for a comparative study of these gliomas in an immunologically intact microenvironment (Amankulor et al., 2017). DF-1 cells with RCAS-PDGF-A + RCAS-hIDHwt-U6-shp53 or RCAS-hIDHmut-U6-shp53 were intracranially injected in nestin-tv-a mice. The second cohort developed tumors with a histological resemblance to human IDH-mutant GBM. Much like human GBM, mice with IDH-mut tumors had longer survival, higher levels of 2-HG, and an increase in overall DNA methylation compared with IDH-wt mice. Strikingly, IDH-mut tumors had less infiltration of various immune cell types (e.g., lymphocytes, macrophages) and impaired neutrophil chemotaxis compared with IDH-wt tumors. These observations could provide some clue for the role of IDH mutations in the glioma microenvironment. Philip et al. (2018) also generated IDH1R132H-driven tumors using the RCAS/tv-a system and found that loss of CDKN2A, ATRX, and PTEN combined with PDGF-A was required to significantly increase tumor penetrance and decrease tumor latency (Philip et al., 2018). These tumors were metabolically, genetically, histologically, and functionally similar to high-grade human IDH-mutant gliomas.
4.4 |. Complexities in studying the glioma immune microenvironment
The glioma microenvironment is a complex ecosystem of tumor cells, immune cells, various brain resident cells, blood vessels, glymphatic structures, and components of the brain ECM (Quail & Joyce, 2017; Sampson, Gunn, Fecci, & Ashley, 2020). The immune landscape of gliomas is characterized by a high density of bone marrow-derived macrophages and resident microglia coupled with low numbers of cytotoxic T-cells. The crosstalk between tumor cells and GAMs drives the creation of a highly immunosuppressive TME via the production of cytokines like IL-6, IL-1β, IL-10, and TGF-β (Kaffes et al., 2019; Szulzewsky et al., 2016), facilitating recruitment of immunosuppressive cells (e.g., myeloid-derived suppressor cells [MDSCs]) and/or transformation to immunosuppressive cellular states that foster immune tolerance and permit tumor growth (Brown, Carter, Ottaviani, & Mulholland, 2018; Quail & Joyce, 2017).
Immunotherapy-based treatments have shown great promise in a wide variety of cancers, revolutionizing the standards of care for many patients (Scheetz et al., 2019; Topalian, Taube, & Pardoll, 2020). Currently, there are several immunotherapies in various stages of preclinical testing and clinical trials as potential treatments for GBM (Boussiotis & Charest, 2018; Sampson et al., 2020). Some of these approaches include boosting the T-cell response against tumor cells via immune checkpoint blockade, tumor vaccines, chimeric antigen receptor T-cell (CAR-T) therapy, or oncolytic virus-based therapies, while others are aimed at reversing the immunosuppressive TME by targeting TAMs and MDSCs (Boussiotis & Charest, 2018; Ene et al., 2020; Lyon, Mokarram, Saxena, Carroll, & Bellamkonda, 2017; Wirsching et al., 2019). For the successful translation of these novel therapies to clinical use, it will be important to select predictive animal models for preclinical testing. Such a model and related host species must possess an immune system bearing a close resemblance to humans and must capture the dynamic interactions that occur between the tumor and its microenvironment beginning during tumor initiation through progression. While it is difficult for a single animal model to emulate all the nuances of the human glioma microenvironment, RCAS-driven GEMs offer certain distinct advantages like in situ tumor formation and gradual evolution of the TME in conjunction with tumor growth (Figure 2) (Olson, Li, Lin, Liu, & Patnaik, 2018; Yeo & Charest, 2017).
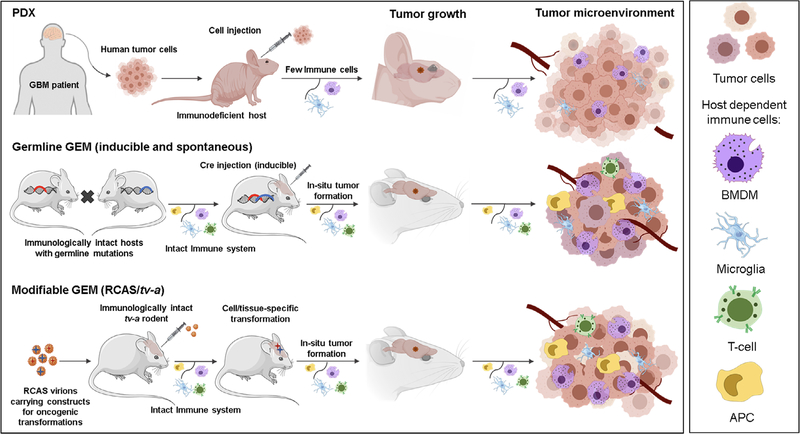
FIGURE 2.
Schematic comparison of PDX, Germline GEM, and Modifiable GEM glioma models. The schematic illustrates the general methodology used to establish the different advanced glioma animal models and details some differences in the major cell types residing in the tumor microenvironments (TME). Notably, all of the models except the Germline GEM (spontaneous) glioma model involve some form of intracranial injection, which may contribute to the tumor initiation and progression process. Human PDX cell lines can be delivered into various immunodeficient rodent strains, ranging from athymic nude (nu) mice (no mature T-cells) to NOD SCID gamma (NSG) mice (no mature T cells, B cells, or natural killer [NK] cells); in each case, the tumor microenvironment will vary depending on the host context. Moreover, modern versions of these PDX models have been set up to restore features of the human TME via an adoptive transfer of human hematopoietic stem cells or differentiated immune cells into immunodeficient rodent hosts creating “humanized” mice. Abbreviations: APC, antigen-presenting cell; BMDM, bone marrow-derived macrophage; GEM, genetically engineered model; PDX, patient-derived xenograft; RCAS, replication-competent ASLV long terminal repeat splice acceptor; tv-a, tumor virus receptor A
RCAS-PDGF-B-driven glioma models have been used to study the composition of the TME in glioma and analyze the expression of inflammatory marker genes in sorted innate immune cells (Bowman et al., 2016; Chen et al., 2017; Szulzewsky et al., 2015). These studies combined the RCAS/tv-a system with fluorescent reporter mice, such as the Cx3cr1GFP/wt-Ccr2RFP/wt knock-in reporter mouse, to distinguish between brain-resident microglia (GFP+) and bone marrow-derived macrophages/monocytes (GFP+/RFP+) within the TME. It was found that GAMs were characterized by a mixed activation state that involved upregulation of both pro-inflammatory (M1-like) and anti-inflammatory or activated (M2-like) genes (Zeiner et al., 2019). Moreover, these expression patterns were different and unique for microglia and the bone marrow-derived macrophages/monocytes (Chen et al., 2017; Szulzewsky et al., 2015). Immunophenotyping based on the dual fluorescence reporter system revealed that infiltrating bone marrow-derived cells (GFP+/RFP+) formed the majority of GAMs in the microenvironment of RCAS-PDGF-B gliomas and localized preferentially to the perivascular areas (Chen et al., 2017; Szulzewsky et al., 2015). Similar findings have been obtained by single-cell RNA sequencing of human GBM samples (Muller et al., 2017). Herting et al. later compared the immune TME of proneural (RCAS-PDGF-B-driven) tumors to the mesenchymal subtype glioma model which is generated by combined Nf1 and p53 silencing. They found that mesenchymal (Nf1-silenced) tumors exhibit an increased presence of Iba1+ microglia/macrophages compared with proneural tumors (Herting et al., 2017), results which were also observed in human mesenchymal and proneural GBM samples (Kaffes et al., 2019). Subsequent studies performing in-depth immunophenotyping of previously identified Iba+ myeloid cell populations in various RCAS-driven GBM subtypes revealed that mesenchymal (Nf1-silenced) tumors possessed a higher percentage of brain-resident microglia while bone marrow-derived macrophages were the major myeloid cell type in the TME of proneural (PDGF-B-driven) and classical (EGFRvIII-mutant) subtypes (Chen et al., 2020), consistent with trends observed in human GBM samples (Gabrusiewicz et al., 2016). Feng et al. showed that loss of Cx3cr1 leads to an increased infiltration of bone marrow-derived monocytes and increased IL-1β production, ultimately leading to a shorter survival of animals bearing RCAS-PDGFB-driven gliomas (Feng et al., 2015). Follow-up studies by Herting et al. showed that pharmacological inhibition of IL-1 signaling could improve the efficacy of radiation therapy of RCAS-PDGF-B tumor-bearing animals (Herting et al., 2019). Aside from immune cells, tumor-associated astrocytes (TAAs) are also found in the microenvironment of proneural gliomas, largely localized to two main regions—the perivascular niche and the invasive tumor margins (Katz et al., 2012). A combination of GFAP and Olig2 fluorescence reporter mice injected with RCAS-PDGF-B reporter constructs were used to determine the distribution and function of these cells. The TAAs have a distinct GBM-associated gene expression profile with increased expression of genes involved in the antigen presentation pathway, among others. Moreover, a subset of these genes had the potential to predict patient survival in g-CIMP proneural tumors.
In another study, Kong et al. described a PDGF-B and Bcl-2-driven glioma model in Ntv-a mice exhibiting key histological features of high-grade gliomas such as diffuse infiltration in the brain parenchyma, pseudopalisading necrosis in brain parenchyma, and migration along white matter tracts (Kong et al., 2010). Immunohistochemical analyses revealed the presence of Tregs and a high percentage of macrophages in these tumors, with the latter negatively impacting the survival of animals with high-grade tumors. These high-grade tumors also had elevated levels of phosphorylated/activated STAT3, an important GBM transcription factor, which correlated with the high degree of macrophage infiltration (Kong et al., 2010). Furthermore, treatment with the STAT3 inhibitor, WP1066, significantly reduced the intratumoral pSTAT3 levels and macrophage numbers resulting in improved overall survival, demonstrating the utility of this model for preclinical testing of targeted therapies in these tumors. Similar RCAS-PDGF mouse models have been used by other groups to study the function of immunosuppressive cell types like MDSCs on the glioma microenvironment. A comparative analysis of tumor samples from one such RCAS-PDGFB mouse model and human GBM patients revealed high levels of MDSCs in both groups (Raychaudhuri et al., 2015). Interestingly, bone marrow-derived MDSCs (BMDCs) were also shown to promote the progression of low-grade RCAS-PDGF-driven lesions to high-grade gliomas in mice as reported by Rajappa et al. (2017). This supported their observation that patients with high-grade gliomas had higher systemic levels of BMDCs than those with low-grade gliomas. Blocking intra-tumoral recruitment of these cells by treatment with a JAK 1/2 inhibitor was shown to be successful in stalling the progression of low-grade lesions to high-grade gliomas in these mice, resulting in improved survival outcomes.
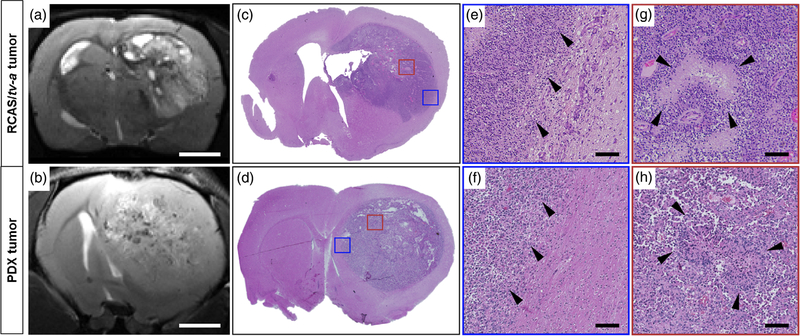
More recently, our group developed a rat glioma model driven by PDGF-A overexpression and p53 knockdown using the RCAS/tv-a system (Connolly et al., 2017). This model faithfully captures many of the classic histological as well as immunological features of human GBM, including few T-cells, microvascular proliferation, regions of pseudopalisading necrosis, and presence of diffusely invasive cells at the tumor margins (Figure 3) (Connolly et al., 2017). Immunofluorescence analysis of the brain tumors showed high macrophage infiltration, the presence of activated microglia, and sparse populations of lymphocytes analogous to human tumors. The distribution of brain resident microglia versus tumor-infiltrating bone marrow-derived macrophages were studied using a CD49d antibody (Bowman et al., 2016). Each of these cell types had distinct patterns of distribution in the tumors and contributed to the composition of the TME. In a subsequent study, the transcriptional profile of these rat tumors was compared with tumors derived from RCAS-PDGF/p53−/− mouse glioma model, spontaneous canine gliomas and human GBM patients (Connolly et al., 2018). Rat and human tumors had the most overlap between core signaling pathways that are critical drivers of glioma biology. On comparing the immune microenvironment signature across tumors, it was observed that only the rat tumors had evidence of high M2-like tumor-promoting macrophage infiltration commonly seen in human tumors.
FIGURE 3.
MRI and histological comparison of RCAS/tv-a and human patient-derived xenograft (PDX) tumors in the rat brain. T2-weighted MRI images reveal some of the complexity and heterogeneity in (a) an RCAS-PDGFA/p53 shRNA-driven tumor and (b) a human GBM39 PDX tumor (Scale bar = 2 mm). Hematoxylin and eosin (H&E) staining of coronal whole brain slices showing the (c) RCAS-PDGFA/p53 shRNA-driven tumor and (d) GBM39 PDX tumor. Colored boxes highlight specific tumor regions for comparison. Blue boxes represent a region of the tumor margin from the (e) RCAS/tv-a and (f) GBM39 PDX tumor. Arrowheads indicate invading tumor cells within adjacent brain regions. (Scale bar = 100 μm). Red boxes represent sections from (g) RCAS/tv-a and (h) GBM39 tumor core regions. Arrowheads highlight areas with pseudopalisading necrosis, a classical feature of human high-grade gliomas. (Scale bar = 100 μm). Abbreviations: PDX, patient-derived xenograft; RCAS, replication-competent ASLV long terminal repeat splice acceptor; tv-a, tumor virus receptor A
4.5 |. Modeling of less common CNS tumors
Several studies have used the RCAS/tv-a system to model less common CNS tumors, such as medulloblastoma and supratentorial ependymomas. Expression of RCAS-SHH (sonic hedgehog protein) alone was sufficient to induce medulloblastoma formation in Ntv-a mice at a low frequency and tumor formation was enhanced substantially by concurrent infection with RCAS-Akt, RCAS-IGF2, or RCAS-N-myc (Browd et al., 2006; Rao et al., 2004). Subsequently, loss of PTEN was later shown to enhance medulloblastoma formation caused by the expression of RCAS-SHH in Ntv-a mice (Hambardzumyan et al., 2008).
Recent large-scale genomic sequencing studies have identified distinct subgroups of Supratentorial (ST)-Ependymoma, characterized by distinct methylation and gene expression patterns and the presence of characteristic gene fusion products that represent the likely tumor-initiating events and oncogenic drivers in these tumors (Lester & McDonald, 2020; Pajtler et al., 2015; Parker et al., 2014). The aggressive ST-EPN-RELA subgroup of ST-ependymoma accounts for ~80–90% of ST-ependymoma cases and is characterized by the presence of the C11orf95-RELA gene fusion (Pajtler et al., 2015; Parker et al., 2014). Using the RCAS/tv-a system to overexpress the most frequent variant of C11orf95-RELA in both Ntv-a and Gtv-a mice, Ozawa et al. have shown that the expression of this gene fusion is sufficient to cause the formation of ependymoma-like tumors that resemble their human counterparts in histomorphology and marker expression (Ozawa et al., 2018). Similar tumors were also observed in Btv-a mice where tv-a expression is driven by brain lipid-binding protein (BLBP) promoter, which is active in the neural stem and progenitor cells present in the ventricular wall (Ozawa et al., 2018). These results indicate that the C11orf95-RELA fusion is the likely tumor-initiating event and oncogenic driver in these tumors and thereby a potential therapeutic target. The less frequent ST-EPN-YAP1 subgroup of ST-ependymoma is characterized by the presence of two distinct YAP1 gene fusions (YAP1-MAMLD1 and YAP1-FAM118B) (Pajtler et al., 2015; Pajtler et al., 2019; Szulzewsky et al., 2020). In addition, YAP1 gene fusions have been identified in several other cancers, where they constitute distinct subgroups of tumor types (Antonescu et al., 2013; Sekine et al., 2019; Sievers et al., 2019). The RCAS/tv-a system was used to express four of these YAP1 gene fusions (YAP1-MAMLD1, YAP1-FAM118B, YAP1-TFE3, and YAP1-SS18) in the brain and hindlimb muscle of Ntv-a or Gtv-a Cdkn2a wild-type and null mice and it was found that expression of each of the analyzed YAP1 fusion genes was sufficient to induce tumor formation in both brain and muscle.
By using the RCAS/tv-a system to express point mutant versions of the different C11orf95-RELA and YAP1 gene fusions, it was possible to examine the necessary amino acid residues for their oncogenic function. Serine 276, located in the Rel homology domain of the RELA protein (S486 in the C11orf95-RELA protein), was identified as essential for the oncogenic functions of C11orf95-RELA and a S486E point mutant C11orf95-RELA was unable to induce tumor formation (Ozawa et al., 2018). Similarly, the interaction between YAP1 and TEAD transcription factors—mediated by the S94 residue of YAP1—was identified as essential for the oncogenic activity of all four analyzed YAP1 fusion proteins and that pharmacological inhibition of this interaction was sufficient to inhibit the growth of RCAS-YAP1-FAM118B-driven mouse tumor cells (Szulzewsky et al., 2020).
4.6 |. Tumor modeling and validation using RCAS/tv-a-CRISPR-Cas9
The RCAS/tv-a system combined with CRISPR-Cas9 genome editing tools offers a powerful additional layer of control in accurately modeling human gliomas. Using engineered RCAS/Nestin-tv-a/Cas9 or RCAS/GFAP-tv-a/Cas9 strains, RCAS-gRNA constructs have been used to edit and knock-out a number of tumor suppressing genes (TSGs) frequently altered in high-grade gliomas (TP53, CDKN2a exon 1β locus, and PTEN) specifically in the sub-ventricular zone, one of the neural stem cell compartments of the brain (Oldrini et al., 2018). Unlike RCAS-PDGFB injections into young or adult Ntv-a and Gtv-a mice, which result in variable tumor penetrance (15–20%) and relatively long latency (>100 days), injection of RCAS-PDGF-B/RCAS-TSG-gRNAs accelerated tumor formation with the majority of tumors having high-grade characteristics and molecular perturbations reflective of the gRNA injected. Single sample gene set enrichment analysis of RNA sequencing data from the RCAS-TSG tumorspheres of these mice showed similarity to the human proneural subtype for Tp53 null cells and the classical subtype in those with both the Cdkn2a and Pten transformations. Importantly, Cas9 induction by tamoxifen exposure did not activate a robust immune response in these adult mice. The RCAS/tv-a-CRISPR-Cas9 system was also used to model and validate recurrent gene fusions as a driver of tumorigenesis using their endogenous promotors (Oldrini et al., 2018). A Bcan-Ntrk1 gene fusion was found to be sufficient to drive glioma formation in NOD/SCID mice. The Bcan-Ntrk1 gliomas were sensitive to the TRK tyrosine kinase inhibitor entrectinib and cells isolated from these gliomas and treated with acute constant doses of entretinib developed resistance. These cells were found to show an increased level of activation of Akt/S6 and MAPK/ERK pathways and showed higher sensitivity to the MAPK inhibitor trametinib as compared with parental Bcan-Ntrk1 tumorspheres, suggesting a novel treatment option for entrectinib-resistant tumors. Using the RCAS/tv-a-CRISPR-Cas9 system, chromosomal translocation and point mutations via homologous recombination were also induced, though in immunocompromised mice. The combined RCAS/tv-a-CRISPR-Cas9 system will enable creative new approaches to study glioma progression and reduce the variability in penetrance and latency which can limit use in preclinical treatment cohorts.
4.7 |. Preclinical testing of therapeutics and imaging modalities
The unique capability of the RCAS/tv-a system to engineer tumors with nearly any desired combination of molecular alterations makes it a highly predictive platform for preclinical testing of therapies targeting specific signaling pathways commonly driving glioma progression (Fomchenko & Holland, 2006; Huse & Holland, 2009; Oldrini et al., 2018) (Table 2). Additionally, these models have also been used to validate the feasibility of new diagnostic imaging technologies for accurate tumor visualization to help with the completeness of surgical resection, optimize currently practiced radiotherapy regimens for better disease management, assess the impact on corticosteroid use on the effectiveness of radiotherapy, and identify molecular drivers of resistance to radiotherapy (Alexander et al., 2020; Karabeber et al., 2014; Leder et al., 2014; Pitter et al., 2016). Given the advantage of using immunocompetent hosts and a TME closely mirroring features of human gliomas, RCAS models have also been useful for exploring the efficacy of novel immunotherapies aimed at boosting antitumor immune responses (Cimino et al., 2019; Ene et al., 2020; Kong et al., 2016; Pyonteck et al., 2013; Quail et al., 2016; Rajappa et al., 2017; Wirsching et al., 2019; Wirsching, Arora, et al., 2019; Zhang et al., 2018). In summary, these studies have helped answer numerous important questions regarding drug development for GBM and shed light on some of the challenges that must be overcome for the successful translation of new therapies to clinical use.
TABLE 2.
Preclinical testing of therapeutics using RCAS/tv-a driven glioma models
| RCAS model | Therapy | Key results | Potential limitations | References |
|---|---|---|---|---|
| RCAS-LacZ, RCAS-K-Ras, RCAS-Akt/HA, RCAS-K-Ras + Akt/HA, and RCAS-PDGF-B | Perifosine (Akt inhibitor) | -Enhanced in vitro and in vivo efficacy when used in combination with temozolomide | (Momota, Nerio, & Holland, 2005) | |
| RCAS-PDGF-B in nestin-tv-a; Ink4a-Arf−/− mice; induced in the brain stem | Perifosine (Akt inhibitor) | -Perifosine in combination with irradiation did not significantly prolong survival compared with irradiation alone | -Tumor model does not significantly reflect the heterogeneity of brain stem gliomas | (Becher et al., 2010) |
| RCAS-PDGF-B + RCAS-Bcl-2 in nestin-tv-a mice | WP1066 (STAT3 inhibitor) | -Oral delivery of WP1066 significantly inhibited intratumoral pSTAT3 levels and recruitment of macrophages -WP1066 treated mice had improved survival |
-Variable rate of tumor incidence and grade may affect randomization for large-scale in vivo studies -Tumor pSTAT3 levels will have to be assessed before treatment |
(Kong et al., 2010) |
| RCAS-PDGF-B, RCAS-IGFBP2 in nestin-tv-a | Mesenchymal stem cells (MSC) | -Syngeneic MSCs are capable of homing to gliomas in immunocompetent mice | -MSC recruitment could be due to tumor overexpression of PDGF-B, potentially making MSC therapy effective only in PDGF-B-driven tumors | (Doucette et al., 2011) |
| RCAS-STAT3 and RCAS-PDGF-B in nestin-tv-a | MicroRNA-124 (miR-124) | -Upregulation of miRNA-124 in glioma cancer stem cells inhibited cancer stem cell immunosuppression -Systemic administration of miRNA-124 and adoptive T-cell therapy showed therapeutic effects |
-Unclear whether what extent of therapeutic effects was mediated by adaptive versus innate immunity | (Wei et al., 2013) |
| RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/− | Radiation | -Mathematical modelingof GBM cell dynamics can predict treatment response and acquisition of radioresistance | -Mathematical model does not include major characteristics of GBM such as the immune system, stomal-tumor interactions, and nutrient gradients | (Leder et al., 2014) |
| RCAS-PDGF-B-RFP in nestin-tv-a; Ink4a-arf−/− Olig2-GFP | Radiation | -Identification of radiation-resistant cell populations that contribute to tumor-recurrence and relapse postradiation treatment | (Alexander et al., 2020) | |
| RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/− | Radiation therapy + dexamethasone | -Corticosteroids that are typically given to patients to alleviate the symptoms of cerebral edema decrease the effectiveness of radiation therapy | (Pitter et al., 2016) | |
| RCAS-PDGF-B in nestin-tv-a | Raman scanner and Raman scattering (SERS) nanoparticles | -Use of a hand-held Raman scanner allow for real-time imaging -Application of SERS nanoparticles significantly enhanced imaging sensitivity |
-Tumor resection studies were performed on brains already fixed with paraformaldehyde | (Karabeber et al., 2014) |
| RCAS-PDGF-B in nestin-tv-a; ink-Arf−/− mice | Sunitinib (multi-kinase inhibitor) | -Oral dosing of sunitinib reduced intra-tumoral and systemic myeloid-derived suppressor cell (MDSC) levels -Increase in overall T-cell levels in tumors also observed |
-No significant survival benefit as a monotherapy; animals eventually succumb to the disease | (Raychaudhuri et al., 2015) |
| Ink4a-Arf−/− ptenloxP/loxP/Ntv-a RCAS/PDGF(+)/Cre(+) | Radiosensitizer gemcitabine, temozolomide, +/− ionizing radiation | -Combination treatments improved survival over single therapies -Diffusion-weighted MRI produced changes in tumor diffusion values before detectable changes in tumor volume |
-Tumor margins were not accurately identifiable potentially introducing error into tumor volume measurements | (Galbán et al., 2012) |
| RCAS-PDGF-B + RCAS-STAT3 in nestin-tv-a mice | Anti-STAT3 miR-124 ± VEGF-4-1BB aptamer conjugate | -miR-124 synergizes with aptamer conjugate to provide a survival benefit -Decreased intra-tumoral GAMMs and increased CD3+ T-cells |
(Kong et al., 2016) | |
| RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl mice | Integrin-directed RGD-SERRS nanoparticles for Raman imaging of GBM tumors | -Integrin targeted RGD-SERRS nanoparticles specifically localized to tumors on systemic delivery -Nanoparticles enabled precise and accurate delineation of invasive tumor margins |
-Raman imaging performed ex vivo on fixed brains | (Huang et al., 2016) |
| RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl mice | Ape1-(DNA-repair enzyme) targeted siRNA nanoparticles + radiotherapy | -Systemic delivery of siRNA-nanoparticles knocks down Ape1 in brain tumors -siRNA nanoparticles mediated Ape1 knockdown synergizes with radiotherapy to extend survival |
-Frequent dosing required to achieve the therapeutic effect -Need to validate efficacy in tumors with different molecular alterations/mutation profile |
(Kievit et al., 2017) |
| RCAS-hPDGF-B-HA + RCAS-Cre in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl mice | AZD1480 (JAK 1/2 inhibitor) | -Oral AZD1480 treatment reduced BMDC recruitment in tumors, stalled progression of low-grade gliomas to high-grade lesions, and prolonged survival | -Indefinite drug administration was required for demonstrating long term survival | (Rajappa et al., 2017) |
|
RCAS-Cre + RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl
EGFRvIII mice |
(PI3K-inhibitor + NKT-cell agonist) liposomes + α-EGFRvIII CAR-T-cells | -Systemic delivery of liposomes before CAR-T cell infusion boosts in vivo CAR-T cell proliferation and antitumor response -liposomes were shown to be nontoxic in multiple doses and can be lyophilized for long-term storage |
(Zhang et al., 2018) | |
| RCAS-PDGF-B + RCAS-KRASG12A in nestin/tv-a; Ink4a/Arf−/− mice | Plinabulin (tubulin inhibitor) | -Plinabulin treatment extended survival of KRAS-driven tumor-bearing mice on systemic delivery | -Intra-tumoral tubulin inhibition was not evaluated in RCAS model -Plinabulin increased pERK levels in vitro in a lung cancer cell line indicating potential modes of resistance |
(Cimino et al., 2019) |
| RCAS-Cre + RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl | ULBP3-encoding HSV-based oncolytic virus (ULBP3-oHSV) + anti PD-1 | -Administration of ULBP3-oHSV alone or with additional anti-PD-1 treatment extended survival | (Wirsching, Zhang, et al., 2019) | |
| RCAS-Cre + RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl | B20 + ULBP3/MMP9-oHSV | -Anti-VEGF therapy with B20/Bevacizumab extends the survival of ULBP3/MMP9-oHSV-treated animals | (Wirsching, Arora, et al., 2019) | |
| RCAS-Cre + RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl with or without EGFRvIII mice | Anti-PD-L1 therapy + radiation therapy (RT) | -Combined anti-PD-L1 and RT extends survival of animals bearing EGFRvIII-expressing tumors | (Ene et al., 2020) | |
| RCAS-Cre + RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl | Anti-CSF-1R therapy (BLZ945) | -Anti-CSF-1R therapy extends survival of animals and results in a shift of macrophage activation toward a more pro-inflammatory phenotype | -Tumors become insensitive to treatment over time | (Pyonteck et al., 2013) |
| RCAS-Cre + RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl | Anti-CSF-1R therapy (BLZ945), anti-PI3Kand anti-IGF-1R blockade | -Combined anti-PI3K and anti-IGF-1R blockade overcomes acquired resistance to anti-CSF-1R therapy | (Quail et al., 2016) | |
| RCAS-Cre + RCAS-PDGF-B in nestin-tv-a; Ink4a-arf−/−/; Ptenfl/fl | Radiation + PbAE-PGA-DiMannose (anti-CD206 moiety) NPs encapsulating IRF5/IKKβ mRNA | -Radiation + NP therapy resulted in more than twofold increase in survival compared with radiation alone | (Zhang et al., 2019) |
Abbreviations: BMDC, bone marrow-derived MDSC; CAR, chimeric antigen receptor; CSF-1R, colony-stimulating factor 1 receptor; EGFRvIII, epidermal growth factor receptor variant III; GAMM, glioma-associated macrophage/microglia; HSV, Herpes simplex virus; IGFBP2, insulin-like growth factor binding protein 2; IGF-1R, insulin growth factor 1 receptor; IKKβ, inhibitor of nuclear factor kappa-B kinase; IRF5, interferon regulatory factor 5; MDSC, myeloid-derived suppressor cell; miR-124, microRNA-124; MMP9, matrix metallopeptidase 9; MRI, magnetic resonance imaging; MSC, mesenchymal stem cell; PbAE, poly-β-amino ester; PDGF, platelet-derived growth factor; PD-L1, programmed death-ligand 1; PGA, poly glutamic acid; RCAS, replication-competent ASLV long terminal repeat with splice acceptor; RT, radiation therapy; SERRS, surface-enhanced resonance Raman spectroscopy; SERS, surface-enhanced Raman scattering; tv-a, tumor virus a; ULBP3, UL-6 binding protein 3; VEGF, vascular endothelial growth factor.
5 |. PERSPECTIVES AND FUTURE DIRECTIONS
The RCAS/tv-a system has proven to be a valuable tool for in vivo modeling of human gliomas. This model system has been especially useful in studying the biology of tumors characterized by single or multiple transforming genetic alterations which function as initiators and/or drivers of tumor progression (e.g., gene fusions, loss of tumor suppressors like Ink4a/Arf, NF-1, etc.) (Antonescu et al., 2013; Ozawa et al., 2018; Szulzewsky et al., 2020; Uhrbom et al., 2002). Similarly, the system is also proving to be quite useful in dissecting the differential effects of alternate splice variants of key genes implicated in driving glioma biology (Pattwell et al., 2020). As we gain a deeper understanding of brain tumor biology and come closer to identifying the triggering mechanism(s) of cellular transformation and cancer initiation, improved cancer diagnostics for earlier detection of and intervention against such changes will be at the forefront. The alternative will be accepting the formidable challenge of treating highly complex, heterogeneous, plastic, and treatment-resistant tumors with some form of therapeutic cocktail that controls for or minimizes the impact of tumor evolution. While not mutually exclusive visions of the future, each vision necessitates a quite different experimental modeling approach and strategy. The RCAS/tv-a modeling system is well-suited for a future where information regarding tumor initiation in one or more specific cell type powers diagnostic and therapeutic strategies to intervene earlier and earlier to effectively manage the disease. Accordingly, work focused on expanding the size and complexity of genomic cassettes, in vivo imaging techniques, inducible constructs, and biological environments to expand and further leverage RCAS/tv-a approaches will be highly valuable. Ongoing examples of this include: examination of neurodevelopmental processes (Beier, Samson, Matsuda, & Cepko, 2011), identifying complex neural circuits in the brain (Stornetta, Inglis, Viar, & Guyenet, 2016; Suzuki, Morimoto, Akaike, & Osakada, 2020), and investigating the developmental origins of other CNS tumors (e.g., medulloblastomas) (Schuller et al., 2008). Controlled genomic manipulations are often essential in dissecting the impact of driver processes and/or biological pathways on disease initiation and progression and validating new treatment strategies. While a single model system will never serve all purposes, genetically engineered RCAS/tv-a models present a powerful tool for advancing future brain cancer research.
ACKNOWLEDGMENTS
Funding to support this work includes National Institutes of Health (NIH) grants K08 NS09043 (G. F. W.), R01 NS107813 (G. F. W.), U54 CA243125-01 (E. C. H.), U54 CA193461 (E. C. H., F. S.). N. P. C. and P. A. were supported in part by NIH Training Grant T32 CA154274. E. C. H. was furthermore supported by the Ivy Foundation Translational Adult Glioma Grant Award from The Ben and Catherine Ivy Foundation.
Funding information
Ben and Catherine Ivy Foundation; National Cancer Institute, Grant/Award Numbers: CA154274, CA193461, CA243125-01; National Institute of Neurological Disorders and Stroke, Grant/Award Numbers: NS09043, NS107813
Footnotes
CONFLICT OF INTEREST
There are no potential conflicts of interest to disclose.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study
REFERENCES
- Agnihotri S, Burrell KE, Wolf A, Jalali S, Hawkins C, Rutka JT, & Zadeh G (2013). Glioblastoma, a brief review of history, molecular genetics, animal models and novel therapeutic strategies. Archivum Immunologiae et Therapiae Experimentalis, 61(1), 25–41. 10.1007/s00005-012-0203-0 [DOI] [PubMed] [Google Scholar]
- Ahronian LG, & Lewis BC (2014). Using the RCAS-TVA system to model human cancer in mice. Cold Spring Harbor Protocols, 2014(11), 1128–1135. 10.1101/pdb.top069831 [DOI] [PubMed] [Google Scholar]
- Alexander J, LaPlant QC, Pattwell SS, Szulzewsky F, Cimino PJ, Caruso FP, … Holland EC (2020). Multimodal single-cell analysis reveals distinct radioresistant stem-like and progenitor cell populations in murine glioma. Glia, 68(12), 2486–2502. 10.1002/glia.23866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amankulor NM, Kim Y, Arora S, Kargl J, Szulzewsky F, Hanke M, … Holland EC (2017). Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes & Development, 31(8), 774–786. 10.1101/gad.294991.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, … Fletcher CD (2013). Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes, Chromosomes & Cancer, 52(8), 775–784. 10.1002/gcc.22073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher OJ, Hambardzumyan D, Walker TR, Helmy K, Nazarian J, Albrecht S, … Holland EC (2010). Preclinical evaluation of radiation and perifosine in a genetically and histologically accurate model of brainstem glioma. Cancer Research, 70(6), 2548–2557. 10.1158/0008-5472.CAN-09-2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher OJ, Hambardzumyan D, Fomchenko EI, Momota H, Mainwaring L, Bleau A-M, …, Holland EC (2008). Gli activity correlates with tumor grade in platelet-derived growth factor–induced gliomas. Cancer Research, 68(7), 2241–2249. 10.1158/0008-5472.can-07-6350 [DOI] [PubMed] [Google Scholar]
- Beier KT, Samson ME, Matsuda T, & Cepko CL (2011). Conditional expression of the TVA receptor allows clonal analysis of descendents from Cre-expressing progenitor cells. Developmental Biology, 353(2), 309–320. 10.1016/j.ydbio.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussiotis VA, & Charest A (2018). Immunotherapies for malignant glioma. Oncogene, 37(9), 1121–1141. 10.1038/s41388-017-0024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RL, Klemm F, Akkari L, Pyonteck SM, Sevenich L, Quail DF, … Joyce JA (2016). Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Reports, 17(9), 2445–2459. 10.1016/j.celrep.2016.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, … Network TR (2013). The somatic genomic landscape of glioblastoma. Cell, 155(2), 462–477. 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg-White JL, Webb CP, Patacsil VS, Miranti CK, Williams BO, & Holmen SL (2004). Delivery of short hairpin RNA sequences by using a replication-competent avian retroviral vector. Journal of Virology, 78(9), 4914–4916. 10.1128/jvi.78.9.4914-4916.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browd SR, Kenney AM, Gottfried ON, Yoon JW, Walterhouse D, Pedone CA, & Fults DW (2006). N-myc can substitute for insulin-like growth factor signaling in a mouse model of sonic hedgehog-induced medulloblastoma. Cancer Research, 66(5), 2666–2672. 10.1158/0008-5472.CAN-05-2198 [DOI] [PubMed] [Google Scholar]
- Brown NF, Carter TJ, Ottaviani D, & Mulholland P (2018). Harnessing the immune system in glioblastoma. British Journal of Cancer, 119(10), 1171–1181. 10.1038/s41416-018-0258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, … Zhang J (2015). Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New England Journal of Medicine, 372(26), 2481–2498. 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Smith DM, Peters MA, Samson ME, Zitz J, Tabin CJ, & Cepko CL (1999). Production and design of more effective avian replication-incompetent retroviral vectors. Developmental Biology, 214(2), 370–384. 10.1006/dbio.1999.9432 [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang Y, Yang J, Hagan JP, & Li M (2013). Vertebrate animal models of glioma: Understanding the mechanisms and developing new therapies. Biochimica et Biophysica Acta, 1836(1), 158–165. 10.1016/j.bbcan.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Feng X, Herting CJ, Garcia VA, Nie K, Pong WW, … Hambardzumyan D (2017). Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Research, 77(9), 2266–2278. 10.1158/0008-5472.CAN-16-2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Herting CJ, Ross JL, Gabanic B, Puigdelloses Vallcorba M., Szulzewsky F, … Hambardzumyan D (2020). Genetic driver mutations introduced in identical cell-of-origin in murine glioblastoma reveal distinct immune landscapes but similar response to checkpoint blockade. Glia, 68(10), 2148–2166. 10.1002/glia.23883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino PJ, Huang L, Du L, Wu Y, Bishop J, Dalsing-Hernandez J, … Wirsching HG (2019). Plinabulin, an inhibitor of tubulin polymerization, targets KRAS signaling through disruption of endosomal recycling. Biomedical Reports, 10(4), 218–224. 10.3892/br.2019.1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloughesy TF, Cavenee WK, & Mischel PS (2014). Glioblastoma: From molecular pathology to targeted treatment. Annual Review of Pathology: Mechanisms of Disease, 9, 1–25. 10.1146/annurev-pathol-011110-130324 [DOI] [PubMed] [Google Scholar]
- Connolly NP, Shetty AC, Stokum JA, Hoeschele I, Siegel MB, Miller CR, … Woodworth GF (2018). Cross-species transcriptional analysis reveals conserved and host-specific neoplastic processes in mammalian glioma. Scientific Reports, 8(1), 1–15. 10.1038/s41598-018-19451-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly NP, Stokum JA, Schneider CS, Ozawa T, Xu S, Galisteo R, … Woodworth GF (2017). Genetically engineered rat gliomas: PDGF-driven tumor initiation and progression in tv-a transgenic rats recreate key features of human brain cancer. PLoS One, 12 (3), e0174557. 10.1371/journal.pone.0174557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, & Holland EC (2001). PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes & Development, 15, 1913–1925. 10.1101/gad.903001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucette T, Rao G, Yang Y, Gumin J, Shinojima N, Bekele BN, … Lang FF (2011). Mesenchymal stem cells display tumor-specific tropism in an RCAS/Ntv-a glioma model. Neoplasia, 13(8), 716–725. 10.1593/neo.101680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, & Li Y (2007). RCAS-TVA in the mammary gland: An in vivo oncogene screen and a high fidelity model for breast transformation? Cell Cycle, 6(7), 823–826. 10.4161/cc.6.7.4074 [DOI] [PubMed] [Google Scholar]
- Ene CI, Kreuser SA, Jung M, Zhang H, Arora S, White Moyes K, … Holland EC (2020). Anti-PD-L1 antibody direct activation of macrophages contributes to a radiation-induced abscopal response in glioblastoma. Neuro-Oncology, 22(5), 639–651. 10.1093/neuonc/noz226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Szulzewsky F, Yerevanian A, Chen Z, Heinzmann D, Rasmussen RD, … Hambardzumyan D (2015). Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget, 6(17), 15077–15094. 10.18632/oncotarget.3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher GH, Orsulic S, Holland E, Hively WP, Li Y, Lewis BC, … Varmus HE (1999). Development of a flexible and specific gene delivery system for production of murine tumor models. Oncogene, 18 (38), 5253–5260. 10.1038/sj.onc.1203087 [DOI] [PubMed] [Google Scholar]
- Fomchenko EI, & Holland EC (2006). Mouse models of brain tumors and their applications in preclinical trials. Clinical Cancer Research, 12 (18), 5288–5297. 10.1158/1078-0432.CCR-06-0438 [DOI] [PubMed] [Google Scholar]
- Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN, … Heimberger AB (2016). Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight, 1(2), e85841. 10.1172/jci.insight.85841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbán S, Lemasson B, Williams TM, Li F, Heist KA, Johnson TD, … Ross BD (2012). DW-MRI as a biomarker to compare therapeutic outcomes in radiotherapy regimens incorporating temozolomide or gemcitabine in glioblastoma. PLoS One, 7(4), e35857–e35857. 10.1371/journal.pone.0035857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressot LV, Doucette TA, Yang Y, Fuller GN, Heimberger AB, Bogler O, … Rao G (2015). Signal transducer and activator of transcription 5b drives malignant progression in a PDGFB-dependent proneural glioma model by suppressing apoptosis. International Journal of Cancer, 136(9), 2047–2054. 10.1002/ijc.29264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday J, Helmy K, Pattwell SS, Pitter KL, LaPlant Q, Ozawa T, & Holland EC (2014). In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proceedings of the National Academy of Sciences of the United States of America, 111(14), 5248–5253. 10.1073/pnas.1321014111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, & Holland EC (2009). Modeling adult gliomas using RCAS/t-va technology. Translational Oncology, 2(2), 89–95. 10.1593/tlo.09100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, & Holland EC (2008). PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes & Development, 22(4), 436–448. 10.1101/gad.1627008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Parada LF, Holland EC, & Charest A (2011). Genetic modeling of gliomas in mice: New tools to tackle old problems. Glia, 59(8), 1155–1168. 10.1002/glia.21142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herting CJ, Chen Z, Maximov V, Duffy A, Szulzewsky F, Shayakhmetov DM, & Hambardzumyan D (2019). Tumour-associated macrophage-derived interleukin-1 mediates glioblastoma-associated cerebral oedema. Brain, 142(12), 3834–3851. 10.1093/brain/awz331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herting CJ, Chen Z, Pitter KL, Szulzewsky F, Kaffes I, Kaluzova M, … Hambardzumyan D (2017). Genetic driver mutations define the expression signature and microenvironmental composition of high-grade gliomas. Glia, 65(12), 1914–1926. 10.1002/glia.23203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, & Fuller GN (2000). Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nature Genetics, 25 (1), 55–57. 10.1038/75596 [DOI] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, & Varmus HE (1998). A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes & Development, 12(23), 3675–3685. 10.1101/gad.12.23.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, & Varmus HE (1998). Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proceedings of the National Academy of Sciences of the United States of America, 95(3), 1218–1223. 10.1073/pnas.95.3.1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KM, Annala M, Chua CY, Dunlap SM, Liu Y, Hugen N, … Zhang W (2012). Insulin-like growth factor-binding protein 2-driven glioma progression is prevented by blocking a clinically significant integrin, integrin-linked kinase, and NF-kappaB network. Proceedings of the National Academy of Sciences of the United States of America, 109(9), 3475–3480. 10.1073/pnas.1120375109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Harmsen S, Samii JM, Karabeber H, Pitter KL, Holland EC, & Kircher MF (2016). High precision imaging of microscopic spread of glioblastoma with a targeted ultrasensitive SERRS molecular imaging probe. Theranostics, 6(8), 1075–1084. 10.7150/thno.13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, … Holland EC (2009). The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes & Development, 23(11), 1327–1337. 10.1101/gad.1777409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, & Holland EC (2009). Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathology, 19(1), 132–143. 10.1111/j.1750-3639.2008.00234.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszthy PC, Daphu I, Niclou SP, Stieber D, Nigro JM, Sakariassen PO, … Bjerkvig R (2012). In vivo models of primary brain tumors: Pitfalls and perspectives. Neuro-Oncology, 14(8), 979–993. 10.1093/neuonc/nos135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffes I, Szulzewsky F, Chen Z, Herting CJ, Gabanic B, Velazquez Vega JE, … Hambardzumyan D (2019). Human mesenchymal glioblastomas are characterized by an increased immune cell presence compared to proneural and classical tumors. Oncoimmunology, 8(11), e1655360. 10.1080/2162402X.2019.1655360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabeber H, Huang R, Iacono P, Samii JM, Pitter K, Holland EC, & Kircher MF (2014). Guiding brain tumor resection using surface-enhanced Raman scattering nanoparticles and a hand-held Raman scanner. ACS Nano, 8(10), 9755–9766. 10.1021/nn503948b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz AM, Amankulor NM, Pitter K, Helmy K, Squatrito M, & Holland EC (2012). Astrocyte-specific expression patterns associated with the PDGF-induced glioma microenvironment. PLoS One, 7 (2), e32453. 10.1371/journal.pone.0032453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kievit FM, Wang K, Ozawa T, Tarudji AW, Silber JR, Holland EC, … Zhang M (2017). Nanoparticle-mediated knockdown of DNA repair sensitizes cells to radiotherapy and extends survival in a genetic mouse model of glioblastoma. Nanomedicine, 13(7), 2131–2139. 10.1016/j.nano.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L-Y, Wei J, Fuller GN, Schrand B, Gabrusiewicz K, Zhou S, … Heimberger AB (2016). Tipping a favorable CNS intratumoral immune response using immune stimulation combined with inhibition of tumor-mediated immune suppression. Oncoimmunology, 5(5), e1117739. 10.1080/2162402X.2015.1117739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LY, Wu AS, Doucette T, Wei J, Priebe W, Fuller GN, … Heimberger AB (2010). Intratumoral mediated immunosuppression is prognostic in genetically engineered murine models of glioma and correlates to immunotherapeutic responses. Clinical Cancer Research, 16(23), 5722–5733. 10.1158/1078-0432.CCR-10-1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe S, Perry A, & Butowski NA (2018). Primary brain tumours in adults. The Lancet, 392(10145), 432–446. 10.1016/s0140-6736(18)30990-5 [DOI] [PubMed] [Google Scholar]
- Leder K, Pitter K, LaPlant Q, Hambardzumyan D, Ross BD, Chan TA, … Michor F (2014). Mathematical modeling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell, 156(3), 603–616. 10.1016/j.cell.2013.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenting K, Verhaak R, Ter Laan M, Wesseling P, & Leenders W (2017). Glioma: Experimental models and reality. Acta Neuropathologica, 133(2), 263–282. 10.1007/s00401-017-1671-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester A, & McDonald KL (2020). Intracranial ependymomas: Molecular insights and translation to treatment. Brain Pathology, 30(1), 3–12. 10.1111/bpa.12781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ferris A, Lewis BC, Orsulic S, Williams BO, Holland EC, & Hughes SH (2012). The RCAS/TVA somatic gene transfer method in modeling human Cancer. In Green J & Ried T (Eds.), Genetically engineered mice for Cancer Research (pp. 83–111). New York, NY: Springer. 10.1007/978-0-387-69805-2_5 [DOI] [Google Scholar]
- Lindberg N, Jiang Y, Xie Y, Bolouri H, Kastemar M, Olofsson T, … Uhrbom L (2014). Oncogenic signaling is dominant to cell of origin and dictates astrocytic or oligodendroglial tumor development from oligodendrocyte precursor cells. Journal of Neuroscience, 34(44), 14644–14651. 10.1523/JNEUROSCI.2977-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg N, Kastemar M, Olofsson T, Smits A, & Uhrbom L (2009). Oligodendrocyte progenitor cells can act as cell of origin for experimental glioma. Oncogene, 28(23), 2266–2275. 10.1038/onc.2009.76 [DOI] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, … Ellison DW (2016). The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathologica, 131(6), 803–820. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- Lyon JG, Mokarram N, Saxena T, Carroll SL, & Bellamkonda RV (2017). Engineering challenges for brain tumor immunotherapy. Advanced Drug Delivery Reviews, 114, 19–32. 10.1016/j.addr.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr U, von Werder A, Seidler B, Reindl W, Bajbouj M, Schmid RM, … Saur D (2008). RCAS-mediated retroviral gene delivery: A versatile tool for the study of gene function in a mouse model of pancreatic cancer. Human Gene Therapy, 19(9), 896–906. 10.1089/hum.2008.014 [DOI] [PubMed] [Google Scholar]
- Misuraca KL, Cordero FJ, & Becher OJ (2015). Pre-clinical models of diffuse intrinsic pontine glioma. Frontiers in Oncology, 5, 172. 10.3389/fonc.2015.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misuraca KL, Hu G, Barton KL, Chung A, & Becher OJ (2016). A novel mouse model of diffuse intrinsic pontine glioma initiated in Pax3-expressing cells. Neoplasia, 18(1), 60–70. 10.1016/j.neo.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyai M, Tomita H, Soeda A, Yano H, Iwama T, & Hara A (2017). Current trends in mouse models of glioblastoma. Journal of Neuro-Oncology, 135(3), 423–432. 10.1007/s11060-017-2626-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momota H, Nerio E, & Holland EC (2005). Perifosine inhibits multiple signaling pathways in glial progenitors and cooperates with temozolomide to arrest cell proliferation in gliomas in vivo. Cancer Research, 65(16), 7429–7435. 10.1158/0008-5472.CAN-05-1042 [DOI] [PubMed] [Google Scholar]
- Muller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, … Diaz A (2017). Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biology, 18(1), 234. 10.1186/s13059-017-1362-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, … Suva ML (2019). An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell, 178(4), 835–849. 10.1016/j.cell.2019.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, … Cancer Genome Atlas Research Network. (2010). Identification of a CpG Island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell, 17(5), 510–522. 10.1016/j.ccr.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh T, Fakurnejad S, Sayegh ET, Clark AJ, Ivan ME, Sun MZ, … Parsa AT (2014). Immunocompetent murine models for the study of glioblastoma immunotherapy. Journal of Translational Medicine, 12, 107. 10.1186/1479-5876-12-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldrini B, Curiel-Garcia A, Marques C, Matia V, Uluckan O, Grana-Castro O, … Squatrito M (2018). Somatic genome editing with the RCAS-TVA-CRISPR-Cas9 system for precision tumor modeling. Nature Communications, 9(1), 1–16. 10.1038/s41467-018-03731-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson B, Li Y, Lin Y, Liu ET, & Patnaik A (2018). Mouse models for cancer immunotherapy research. Cancer Discovery, 8(11), 1358–1365. 10.1158/2159-8290.CD-18-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omuro A, & DeAngelis LM (2013). Glioblastoma and other malignant gliomas: A clinical review. JAMA, 310(17), 1842–1850. 10.1001/jama.2013.280319 [DOI] [PubMed] [Google Scholar]
- Orsulic S (2002). An RCAS-TVA-based approach to designer mouse models. Mammalian Genome, 13(10), 543–547. 10.1007/s00335-002-4003-4 [DOI] [PubMed] [Google Scholar]
- Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, & Barnholtz-Sloan JS (2020). CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro-Oncology, 22(S1), v1–v96. 10.1093/neuonc/noaa200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Arora S, Szulzewsky F, Juric-Sekhar G, Miyajima Y, Bolouri H, … Holland EC (2018). A de novo mouse model of C11orf95-RELA fusion-driven ependymoma identifies driver functions in addition to NF-kappaB. Cell Reports, 23(13), 3787–3797. 10.1016/j.celrep.2018.04.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, … Holland EC (2014). Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell, 26 (2), 288–300. 10.1016/j.ccr.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajtler KW, Wei Y, Okonechnikov K, Silva PBG, Vouri M, Zhang L, … Kawauchi D (2019). YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nature Communications, 10(1), 1–16. 10.1038/s41467-019-11884-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, … Pfister SM (2015). Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell, 27(5), 728–743. 10.1016/j.ccell.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, … Gilbertson RJ (2014). C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature, 506(7489), 451–455. 10.1038/nature13109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattwell SS, Arora S, Cimino PJ, Ozawa T, Szulzewsky F, Hoellerbauer P, … Holland EC (2020). A kinase-deficient NTRK2 splice variant predominates in glioma and amplifies several oncogenic signaling pathways. Nature Communications, 11(1), 1–14. 10.1038/s41467-020-16786-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip B, Yu DX, Silvis MR, Shin CH, Robinson JP, Robinson GL, … Holmen SL (2018). Mutant IDH1 promotes glioma formation in vivo. Cell Reports, 23(5), 1553–1564. 10.1016/j.celrep.2018.03.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirozzi CJ, Carpenter AB, Waitkus MS, Wang CY, Zhu H, Hansen LJ, … Yan H (2017). Mutant IDH1 disrupts the mouse subventricular zone and alters brain tumor progression. Molecular Cancer Research, 15(5), 507–520. 10.1158/1541-7786.MCR-16-0485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitter KL, Tamagno I, Alikhanyan K, Hosni-Ahmed A, Pattwell SS, Donnola S, … Hambardzumyan D (2016). Corticosteroids compromise survival in glioblastoma. Brain, 139(Pt 5), 1458–1471. 10.1093/brain/aww046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, … Joyce JA (2013). CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nature Medicine, 19(10), 1264–1272. 10.1038/nm.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, Bowman RL, Akkari L, Quick ML, Schuhmacher AJ, Huse JT, … Joyce JA (2016). The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science, 352 (6288), aad3018. 10.1126/science.aad3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, & Joyce JA (2017). The microenvironmental landscape of brain tumors. Cancer Cell, 31(3), 326–341. 10.1016/j.ccell.2017.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajappa P, Cobb WS, Vartanian E, Huang Y, Daly L, Hoffman C, … Greenfield JP (2017). Malignant astrocytic tumor progression potentiated by JAK-mediated recruitment of myeloid cells. Clinical Cancer Research, 23(12), 3109–3119. 10.1158/1078-0432.CCR-16-1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao G, Pedone CA, Del Valle L, Reiss K, Holland EC, & Fults DW (2004). Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene, 23(36), 6156–6162. 10.1038/sj.onc.1207818 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri B, Rayman P, Huang P, Grabowski M, Hambardzumyan D, Finke JH, & Vogelbaum MA (2015). Myeloid derived suppressor cell infiltration of murine and human gliomas is associated with reduction of tumor infiltrating lymphocytes. Journal of Neuro-Oncology, 122(2), 293–301. 10.1007/s11060-015-1720-6 [DOI] [PubMed] [Google Scholar]
- Robinson JP, VanBrocklin MW, Guilbeault AR, Signorelli DL, Brandner S, & Holmen SL (2010). Activated BRAF induces gliomas in mice when combined with Ink4a/Arf loss or Akt activation. Oncogene, 29(3), 335–344. 10.1038/onc.2009.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JH, Gunn MD, Fecci PE, & Ashley DM (2020). Brain immunology and immunotherapy in brain tumours. Nature Reviews Cancer, 20(1), 12–25. 10.1038/s41568-019-0224-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, & Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America, 106(33), 13939–13944. 10.1073/pnas.0907143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz L, Park KS, Li Q, Lowenstein PR, Castro MG, Schwendeman A, & Moon JJ (2019). Engineering patient-specific cancer immunotherapies. Nature Biomedical Engineering, 3(10), 768–782. 10.1038/s41551-019-0436-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, … Ligon KL (2008). Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell, 14(2), 123–134. 10.1016/j.ccr.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler B, Schmidt A, Mayr U, Nakhai H, Schmid RM, Schneider G, & Saur D (2008). A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proceedings of the National Academy of Sciences of the United States of America, 105(29), 10137–10142. 10.1073/pnas.0800487105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Kiyono T, Ryo E, Ogawa R, Wakai S, Ichikawa H, … Mori T (2019). Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. Journal of Clinical Investigation, 129(9), 3827–3832. 10.1172/JCI126185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, & Holland EC (2004). Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Research, 64(14), 4783–4789. 10.1158/0008-5472.CAN-03-3831 [DOI] [PubMed] [Google Scholar]
- Sievers P, Chiang J, Schrimpf D, Stichel D, Paramasivam N, Sill M, … Sahm F (2019). YAP1-fusions in pediatric NF2-wildtype meningioma. Acta Neuropathologica, 139, 215–218. 10.1007/s00401-019-02095-9 [DOI] [PubMed] [Google Scholar]
- Stornetta RL, Inglis MA, Viar KE, & Guyenet PG (2016). Afferent and efferent connections of C1 cells with spinal cord or hypothalamic projections in mice. Brain Structure and Function, 221(8), 4027–4044. 10.1007/s00429-015-1143-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylli SS, Luwor RB, Ware TM, Tan F, & Kaye AH (2015). Mouse models of glioma. Journal of Clinical Neuroscience, 22(4), 619–626. 10.1016/j.jocn.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Morimoto N, Akaike A, & Osakada F (2020). Multiplex neural circuit tracing with G-deleted rabies viral vectors. Frontiers in Neural Circuits, 13, 77. 10.3389/fncir.2019.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulzewsky F, Arora S, de Witte L, Ulas T, Markovic D, Schultze JL, … Kettenmann H (2016). Human glioblastoma-associated microglia/monocytes express a distinct RNA profile compared to human control and murine samples. Glia, 64(8), 1416–1436. 10.1002/glia.23014 [DOI] [PubMed] [Google Scholar]
- Szulzewsky F, Arora S, Hoellerbauer P, King C, Nathan E, Chan M, … Holland EC (2020). Comparison of tumor-associated YAP1 fusions identifies a recurrent set of functions critical for oncogenesis. Genes & Development, 34(15–16), 1051–1064. 10.1101/gad.338681.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulzewsky F, Pelz A, Feng X, Synowitz M, Markovic D, Langmann T, … Kettenmann H (2015). Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One, 10(2), e0116644. 10.1371/journal.pone.0116644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Taube JM, & Pardoll DM (2020). Neoadjuvant checkpoint blockade for cancer immunotherapy. Science, 367(6477), eaax0182. 10.1126/science.aax0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrbom L, Dai C, Celestino JC, Rosenblum MK, Fuller GN, & Holland EC (2002). Ink4a-arf loss cooperates with Kras activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Research, 62(19), 5551–5558. [PubMed] [Google Scholar]
- Uhrbom L, Nerio E, & Holland EC (2004). Dissecting tumor maintenance requirements using bioluminescence imaging of cell proliferation in a mouse glioma model. Nature Medicine, 10(11), 1257–1260. 10.1038/nm1120 [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, … Cancer Genome Atlas Research N (2010). Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell, 17(1), 98–110. 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE (2011). Cells of origin in cancer. Nature, 469(7330), 314–322. 10.1038/nature09781 [DOI] [PubMed] [Google Scholar]
- Von Werder A, Seidler B, Schmid RM, Schneider G, & Saur D (2012). Production of avian retroviruses and tissue-specific somatic retroviral gene transfer in vivo using the RCAS/TVA system. Nature Protocols, 7 (6), 1167–1183. 10.1038/nprot.2012.060 [DOI] [PubMed] [Google Scholar]
- Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, … Verhaak RGW (2017). Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell, 32(1), 42–56. 10.1016/j.ccell.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, … Heimberger AB (2013). MiR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Research, 73(13), 3913–3926. 10.1158/0008-5472.CAN-12-4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen PY, & Kesari S (2008). Malignant gliomas in adults. New England Journal of Medicine, 359(5), 492–507. 10.1056/NEJMra0708126 [DOI] [PubMed] [Google Scholar]
- Wesseling P, & Capper D (2018). WHO 2016 classification of gliomas. Neuropathology and Applied Neurobiology, 44(2), 139–150. 10.1111/nan.12432 [DOI] [PubMed] [Google Scholar]
- Wirsching HG, Arora S, Zhang H, Szulzewsky F, Cimino PJ, Queva C, … Holland EC (2019). Cooperation of oncolytic virotherapy with VEGF-neutralizing antibody treatment in IDH wildtype glioblastoma depends on MMP9. Neuro-Oncology, 21(12), 1607–1609. 10.1093/neuonc/noz145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsching HG, Zhang H, Szulzewsky F, Arora S, Grandi P, Cimino PJ, … Holland EC (2019). Arming oHSV with ULBP3 drives abscopal immunity in lymphocyte-depleted glioblastoma. JCI Insight, 4(13), e128217. 10.1172/jci.insight.128217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, … Bigner DD (2009). IDH1 and IDH2 mutations in gliomas. New England Journal of Medicine, 360(8), 765–773. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo AT, & Charest A (2017). Immune checkpoint blockade biology in mouse models of glioblastoma. Journal of Cellular Biochemistry, 118(9), 2516–2527. 10.1002/jcb.25948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner PS, Preusse C, Golebiewska A, Zinke J, Iriondo A, Muller A, … Mittelbronn M (2019). Distribution and prognostic impact of microglia/macrophage subpopulations in gliomas. Brain Pathology, 29(4), 513–529. 10.1111/bpa.12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Parayath NN, Ene CI, Stephan SB, Koehne AL, Coon ME, … Stephan MT (2019). Genetic programming of macrophages to perform anti-tumor functions using targeted mRNA nanocarriers. Nature Communications, 10(1), 1–16. 10.1038/s41467-019-11911-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Stephan SB, Ene CI, Smith TT, Holland EC, & Stephan MT (2018). Nanoparticles that reshape the tumor milieu create a therapeutic window for effective T-cell therapy in solid malignancies. Cancer Research, 78(13), 3718–3730. 10.1158/0008-5472.CAN-18-0306 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study