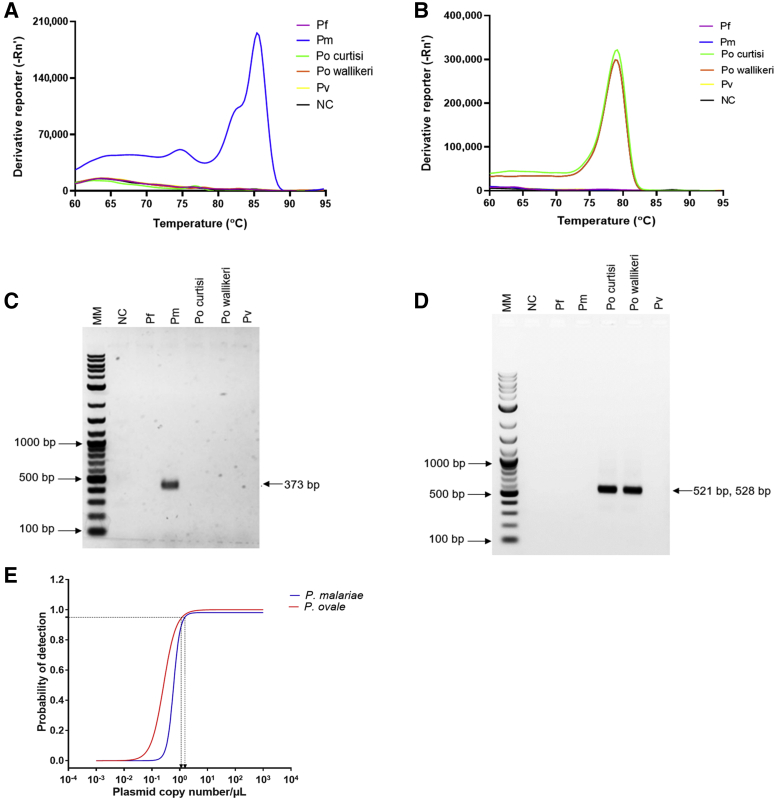
Figure 1.
The specificity and detection limits of Plasmodium malariae and Plasmodium ovale assays. Assays were performed using Plasmodium falciparum (Pf), P. malariae (Pm), P. ovale curtisi (Po curtisi), P. ovale wallikeri (Po wallikeri), and Plasmodium vivax (Pv) genomic DNA. A: Melt curve for P. malariae (blue). B: Melt curves for P. ovale curtisi (green) and P. ovale wallikeri (orange) assays. Other colors represent the P. falciparum, P. vivax, and nontemplate control (NC). C: Separation of the resulting P. malariae real-time quantitative PCR (qPCR) amplicons on 1.5% agarose gel. D: Separation of the resulting qPCR amplicons for P. ovale subspecies on 1.5% agarose gel. Expected amplicon sizes for P. ovale curtisi and P. ovale wallikeri were 528 and 521 bp, respectively. E: The detection limits for P. malariae (blue curve) and P. ovale (red curve) assays were determined using a 10-fold serial dilution of plasmids. Plasmid concentrations were log-transformed and then analyzed using probit analysis. The probability of detection was plotted against the plasmid copy numbers/μL of the DNA. Molecular weight marker (MM) shown in bp.