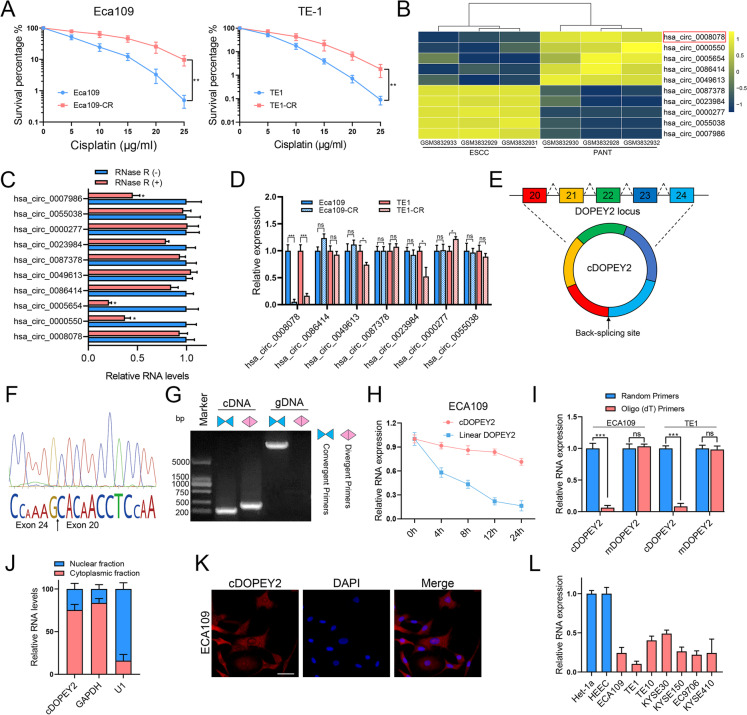
Fig. 1.
Identification and characterization of a cisplatin sensitivity-associated circRNA (cDOPEY2). A The cisplatin sensitivity of two cisplatin-resistant ESCC cell lines (ECA109-CR and TE1-CR) was determined by a clonogenic formation assay. B A heatmap showing the top 5 most upregulated circRNAs and 5 most downregulated circRNAs in ESCC tissues and paired adjacent nontumorous tissues (PANTs) in GSE131969. cDOPEY2 (hsa_circ_0008078) is indicated by the red box. C qPCR analysis of the RNA levels of the indicated circRNAs with or without RNase R treatment. D qPCR analysis of the RNA levels of the indicated circRNAs in cisplatin-resistant cells and their corresponding parental cells. E A schematic diagram showing that cDOPEY2 was formed by the back-splicing of linear cDOPEY2 between the 20th and 24th exons. F The cDOPEY2 junction site was identified by Sanger sequencing. G qPCR analysis of cDOPEY2 and its linear counterpart DOPEY2 in complementary DNA (cDNA) and genomic DNA (gDNA). H Time-course qPCR analysis of cDOPEY2 and its linear counterpart DOPEY2 in ECA109 cells treated with actinomycin D (5 µg/mL). I qPCR amplification of cDOPEY2 and its linear counterpart DOPEY2 using random hexamer primers and oligo (dT) primers. J Subcellular qPCR analysis showing that cDOPEY2 was mainly localized in the cytoplasm. GAPDH and U1 were applied as positive controls in the cytoplasm and nucleus, respectively. K RNA fluorescence in situ hybridization analysis revealed the subcellular localization of cDOPEY2. Scale bar, 20 μm. L Relative expression of cDOPEY2 in a set of ESCC cell lines and normal esophageal epithelial cells (Het-1a and HEEC). Data are presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. P values were determined using the unpaired Student’s t-test (A, C, D, H and I)