Figure 3. M13-mediated delivery of CRISPR-Cas9 to E. coli in vitro causes impaired colony growth and can induce chromosomal deletions that encompass the targeted gene.
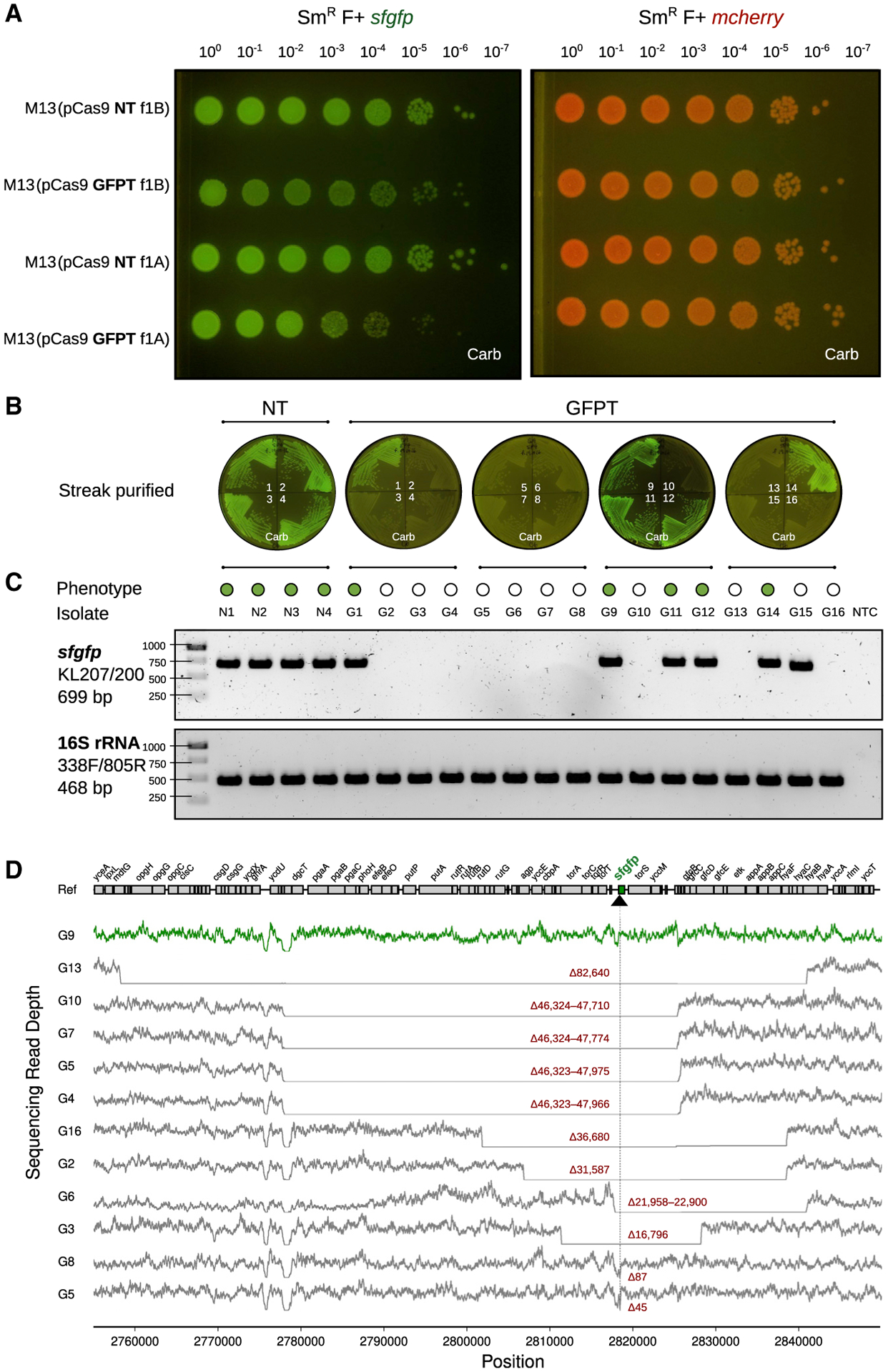
(A)GFP+E. coliexhibit a sick colony morphologyafter infection with M13 phage carrying GFP-targeting (GFPT) CRISPR-Cas9. NT (non-targeting) or GFPT M13 were used to infect SmR W1655 F+ sfgfp or SmR W1655 F+ mcherry as a control. Cells were infected, diluted, and spotted onto media with selection for the vector; f1A or f1B indicates vector version.
(B) CRISPR-Cas9 targeting the sfgfp gene can induce loss of fluorescence. Colonies arising from infection with NT-M13 or GFPT-M13 were subjected to several rounds of streak purification on selective media to ensure phenotypic homogeneity and clonality. The majority (11/16) of GFPT clones exhibited a loss of fluorescence.
(C) Clones exhibiting loss of fluorescence either lack an sfgfp PCR amplicon or exhibit an amplicon of decreased size. Genomic DNA was isolated from streak-purified clones, and PCR was used to determine whether the sfgfp gene was present. PCR for the 16S rRNA gene was performed as a positive control.
(D) Genome-sequencing results confirm that non-fluorescent clones have chromosomal deletions encompassing the targeted gene. Read depth surrounding sfgfp locus for a fluorescent control clone G9 (green line) and all non-fluorescent clones (gray lines). Deletion size is indicated in red; range indicates a deletion flanked by repetitive sequences. Black arrow and vertical line denote position of targeting. See also Figure S4.
