Figure 7. Mechanisms enabling GFP+ mCherry+ E. coli to escape CRISPR-Cas9 targeting.
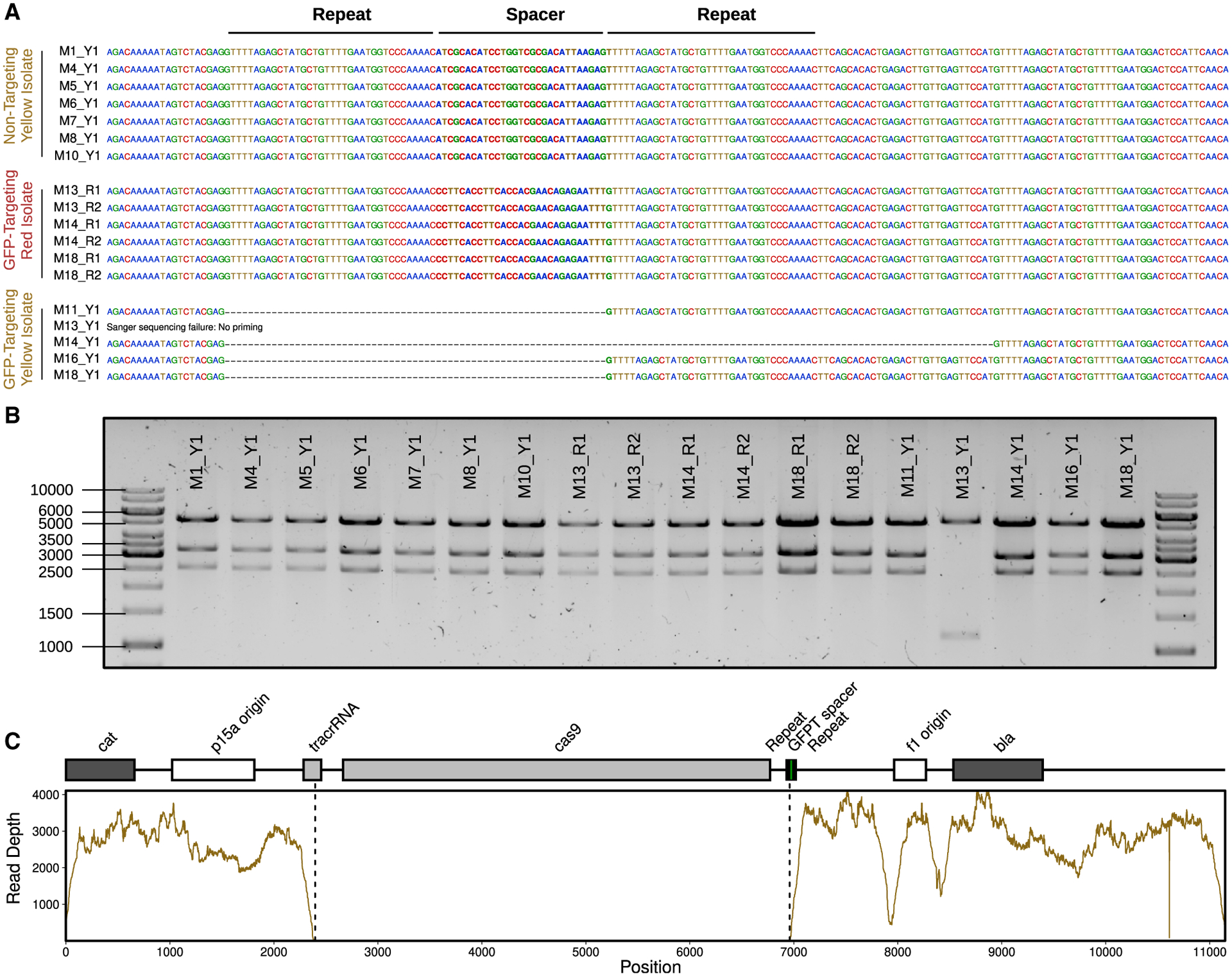
(A) Sanger-sequencing results confirm the expected spacer present in phagemid DNA extracted from fluorescent yellow isolates (Y1) colonizing NT mice (M1, M4, M5, M6, M7, M8, and M10) and fluorescent red isolates (R1 and R2) colonizing GFPT mice (M13, M14, and M18). In contrast, 4 out of 5 fluorescent yellow isolates colonizing GFPT mice (M11, M13, M14, M16, and M18) were confirmed to have lost the spacer. No Sanger-sequence data were obtained for the last isolate (M13) with the report for failing being “no priming,” suggesting loss of a larger fragment from the phagemid.
(B) Diagnostic digest of CRISPR-Cas9 phagemid DNA indicates loss of a portion of phagemid DNA for the phagemid extracted from M13 Y1. Expected fragment sizes from KpnI-XbaI double digest: 5,289, 3,285, and 2,573 bp.
(C) Genome-sequencing data for M13 Y1 confirms loss of DNA from phagemid. Sequencing coverage across the GFPT phagemid reveals lack of reads corresponding to the cas9 gene and parts of the CRISPR array and tracrRNA.
