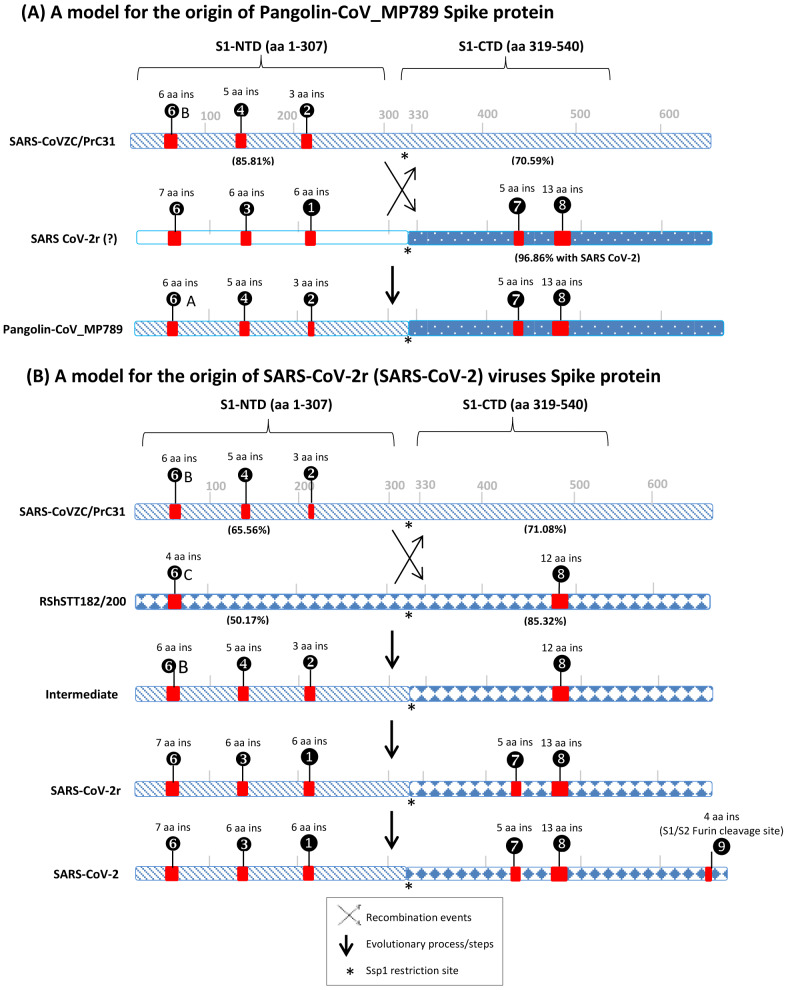
Figure 6. A conceptual diagram (models) summarizing the sequence characteristics of the spike protein from specific lineages of sarbecoviruses and their implications regarding the origin of Pangolin-CoV_MP789 virus and SARS-CoV-2r cluster of viruses.
(A) Sequence characteristics of the S1-NTD and S1-CTD from SARS-CoVZC45, a SARS-CoV-2r virus and pangolin-CoV_MP789 and a model based upon them for the origin of the pangolin-CoV_MP789 virus. The ? mark besides the SARS-CoV-2r virus indicates that the virus involved in this recombination remains unidentified. The percent identity (96.86%) shown here is for the Wuhan-Hu-1 virus. The CSIs that are present (or predicted to be present) in these sequence regions and their sequence lengths are noted above the lines. The % amino acid sequence identity noted on the top line is to the indicated region to the pangolin-CoV_MP789 virus. The cross (x) in (A) and (B) indicates a genetic recombination event, which is postulated to have occurred at (near) the S1-NTD and S1-CTD boundary, where a Ssp1 site (marked by *) is present (B) The sequence characteristics and distribution pattern of different CSIs in the S1-NTD and S1-CTD of SARS-CoVZC45, RShSTT182/200 and SARS-CoV-2r cluster of CoVs and a model based on them for the origin of the SARS-CoV-2r cluster of viruses. The numbers below the lines in parenthesis indicate the % amino acid sequence identity of the indicated regions to the SARS-CoV-2 Wuhan-Hu-1 virus. The intermediate is a postulated stage resulting from recombination, prior to the occurrence of further evolutionary changes in it. The vertical arrows indicate other genetic changes in the evolution of SARS-CoV-2r virus including acquisition of the 4 aa insertion (❾) by SARS-CoV-2 at the S1 and S2 boundary. The * indicates the presence of a Ssp1 restriction site in the sequences of these viruses, which is indicated to be the presumed site of recombination.