Figure 4.
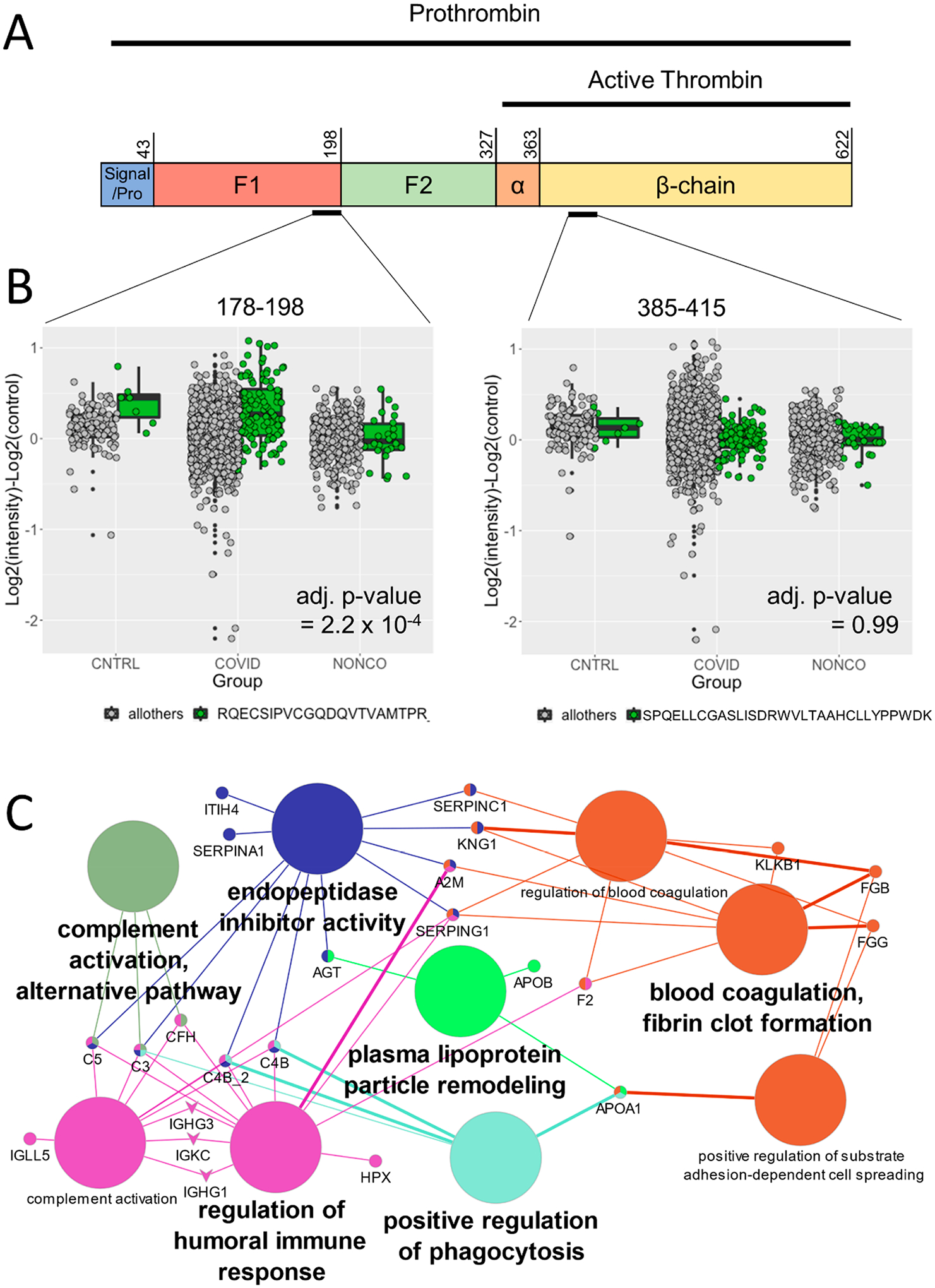
PeCorA detects altered plasma proteoforms associated with COVID-19 infection. (A) Schematic representation of the prothrombin primary sequence, which is the precursor of active thrombin. Prothrombin contains N-terminal pro and signal sequences (residues 1–43) followed by two fragment peptides that must be removed to activate thrombin, F1 (residues 44–198) and F2 (residues 199–327). The active protease domain comprises residues 328–622. Once in its active form, thrombin promotes coagulation. (B) Left: Prothrombin peptide covering residues 177–198 of the inactive F1 region identified as significantly elevated by PeCorA in COVID-19 ARDS patients relative to non-COVID-19 ARDS patients. Right: Unchanged thrombin peptide from the active β-chain 385–415. (C) Enriched GO Biological Process terms from the 26 proteins with altered proteoforms based on the PeCorA adjusted p value < 0.01. The big circles are the GO Biological Process terms, the small circles are the proteins, and the edges show how the proteins are members of the various GO terms. GO terms were filtered to show only the minimal subset needed to make all of the connections between proteins.
