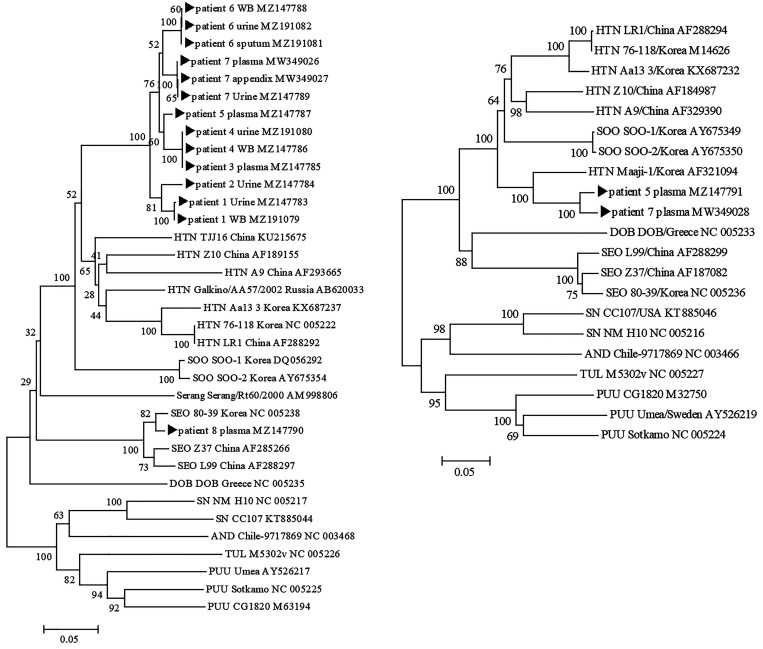
Figure 1.
Phylogenetic trees for hantaviruses based on the partial large (L)-segment genome sequences (360 nt) (A) and based on the partial small (S)-segment genome sequences (645 nt) (B). The CLUSTAL X software program (http://www.clustal.org/clustal2/) was used to construct the phylogenetic trees by using neighbor-joining (NJ) with 1,000 bootstrap replicates. HTN = Hantaan virus; SEO = Seoul virus; SOO = Soochong virus; SN = sin nombre virus; PUU = Puumala virus; AND = Andes virus; TUL = Tula virus; DOB = Dobrava-Belgrade virus.