Abstract
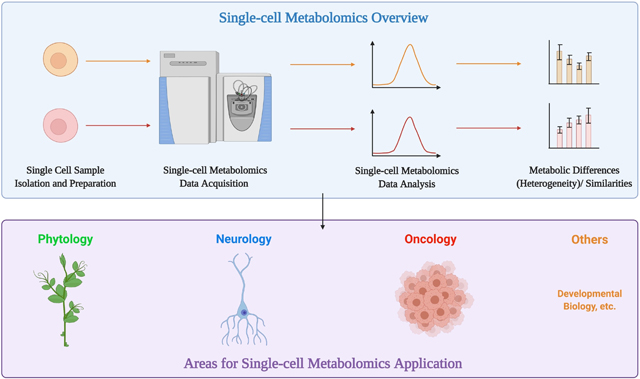
Single-cell metabolomics (SCM) is currently one of the most powerful tools for performing high-throughput metabolic analysis at the cellular level. The differences among individual cells in biological samples have posed significant challenges. Therefore, single-cell metabolomics’ power in determining metabolic profiles for individual cells makes it very suitable to decode cell heterogeneity. SCM bears great potential in cell type identification and differentiation within cell colonies. With the development of various equipment and techniques, single-cell metabolic analysis has become available for a wide range of biological samples. Many fields have incorporated this cutting-edge analytic tool and generated fruitful findings. This review article pays close attention to the prevalent techniques utilized in SCM and the exciting new findings and applications developed by studies in phytology, neurology, and oncology using SCM.
Keywords: Single-cell metabolomics, mass spectrometry, cell heterogeneity, oncology, neurology, phytology
Graphical Abstract
Introduction
The metabolome refers to all small-molecule metabolites within a biological sample, from a single cell to the whole organism [1]. These metabolites are crucial components of most intracellular reactions and functions, and reflect the given sample’s physiological state. Therefore, quantitative metabolomics has long been a powerful analytic tool in many fields. Metabolomic analysis has generated exciting findings [2–4]. Due to sensitivity factors, current metabolomics methods usually require a large number of cells for each sample, especially for some methods such as nuclear magnetic resonance (NMR) [5]. Although multi-cell samples allow for adequate detection of chemical components, the heterogeneity of cells is overlooked. The differences among individual cells may arise as a result of genetic, epigenetic, developmental, and environmental factors. Even cells in a clonal or isogenic culture may differ in phenotype due to stochastic biological processes [5]. Recent single-cell genetics and proteomics studies provide information regarding tumor heterogeneity that was overlooked by previous studies on cell populations [6,7]. Therefore, SCM, which can reveal the cellular metabolome’s differences among individual cells, is in great demand.
Detection and identification of specific analytes at the single-cell level have long been available. However, SCM further expands the analytes range and enables their effective analysis [8,9]. It helps gather information on the chemical components and metabolic fluxes in a single cell, leading to further understanding of cell functions [9]. Several studies have shown promising results in producing metabolic data from different cell and tissue types [10–12]. This review article will focus on the high-potential mass spectrometry (MS)-based technologies emerging in SCM and the exciting, novel findings resulting from their development.
1. Current Methods of Single Cell Metabolic Analysis
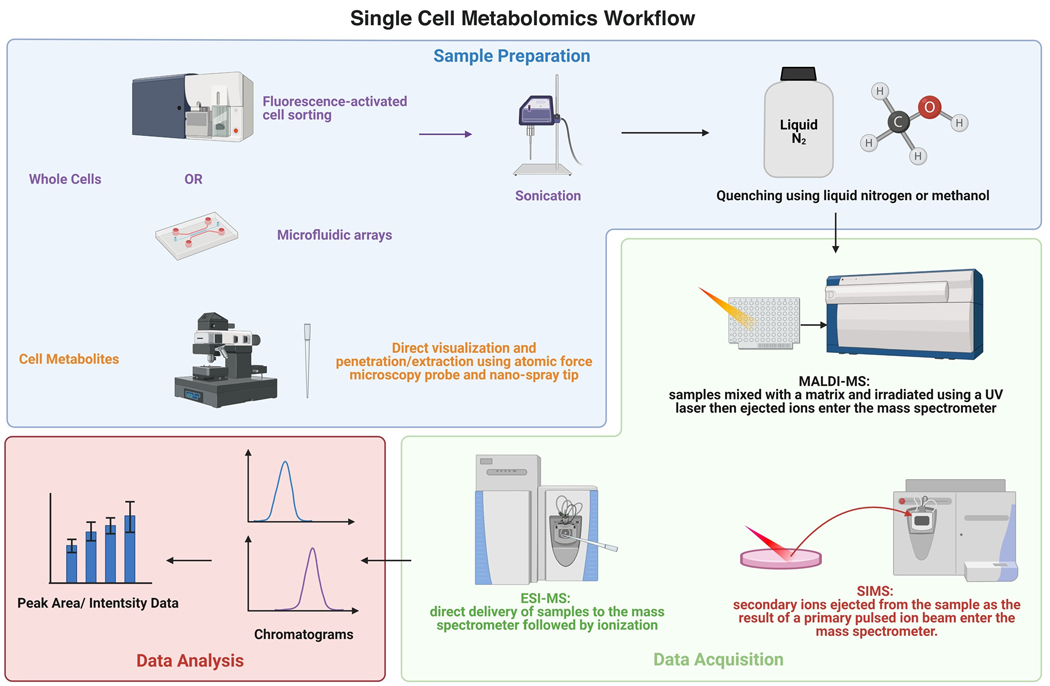
A typical workflow of SCM starts with sample preparation to isolate a single cell or its cell content (Figure 1). Common methods of sample preparation include fluorescence-activated cell sorting (FACS), microfluidic arrays, and direct visualization and penetration/extraction via an atomic force microscopy (AFM) probe [1,13]. The first two methods preserve the original morphology of whole cells, while the last strategy only keeps the cell metabolites in the probe. For whole-cell samples, sonication is reported to increase detection sensitivity, while direct delivery from the probe to the mass spectrometer allows minimal disruption [14]. A common setup includes stereomicroscopes, nano-spray tips connected to a syringe, and a micromanipulator, which allows for both visualization and precise control of extraction [15,16].
Fig 1.
Single Cell Metabolomics’ Workflow. Created with BioRender.com
Immediately after isolating a single cell, the cellular metabolism should be quenched using an organic acid or primary solvent to denature enzymes and prevent further conversion of metabolites to products. One of the most common quenching methods is snap freezing using liquid nitrogen to stop cellular metabolism and promote cell lysis [17]. Freeze-quenching is easy to perform and is compatible with most MS methods as it is free of added reagents. Organic solvents are also commonly used for cellular metabolism quenching. The advantage of this method is that the organic solvents used can also help with metabolites extraction. For example, acetonitrile is used for both quenching and extraction for matrix-assisted laser desorption/ionization MS (MALDI-MS) [18]. One of the most commonly used organic solvents for quenching is methanol. While the pre-cooled methanol/water mixture is most widely used, it is also often used in combination with other organic or inorganic reagents as needed, such as saline, ammonium carbonate, and HEPES [19]. A combination of 60% methanol and 0.85% ammonium bicarbonate (AMBIC) has also been proposed as an effective method for mammalian cell quenching [19,20]. A study done by Chen et al. compared the effect of 60% and 80% methanol/water mixture and 80% methanol/glycerol mixture on quenching L. bulgaricus for metabolic extraction and found the traditional methanol/water combination exhibited the least metabolite leakage [21]. Therefore, methanol-based quenching methods stand out to be the most prevalent and effective quenching strategies so far.
Among all methods in SCM, MS-based technologies stand out above the rest. They do not require labeling and are highly sensitive. With the development of nano-scale matrices, MALDI-MS has become one of the most popular techniques used for single-cell metabolic analysis. To perform MALDI-MS measurements, the samples are first mixed with a matrix followed by irradiation under vacuum using a UV laser beam. The ionized and accelerated analytes then enter the mass spectrometer [22]. The high throughput of MALDI-MS enables the classification of cellular subtypes and the detection of individual cells within a large cell population, as reported by Ong et al. [23]. MALDI-MS has also been used to reveal the intrapopulation heterogeneity of unicellular organisms [24].
Electrospray ionization MS (ESI-MS) is another technique widely used in single-cell metabolic analysis. It usually involves direct delivery of samples from a probe or capillary to the mass spectrometer with minimal pretreatment. Due to ionization under ambient conditions, ESI-MS prevents sample disruption. Therefore, it is widely used in live single-cell metabolic analysis. However, its limitations include relatively low resolution and low throughput, making it not suitable for system biology research [25]. Progress has been made to address these two limitations. ESI-MS can be used in conjunction with capillary electrophoresis (CE) [26]. A recent study by Kawai et al. discusses how they significantly improved CE-MS resolution using a self-designed ionization emitter [27]. Another study integrates droplet-based microextraction with ESI-MS and successfully prevents matrix interference while ensuring a high throughput [28]. Therefore, ESI-MS has a great potential to become more widely used in single-cell metabolic research.
Secondary ion MS (SIMS) is a relatively new technique used in SCM with super high resolution (<100 nm to 1 μm) [1]. For SIMS, a primary pulsed ion beam is shot onto the sample’s surface, and the ejected secondary ions are measured using a mass spectrometer. Its ultrahigh-resolution makes it suitable for analyzing metabolites in subcellular spaces [22]. However, it could also cause damage to larger organic molecules due to the strong primary ion dose [1]. Additionally, SIMS does not support full-scan and MS/MS analyses simultaneously and has low throughput, though it can be improved by sacrificing the resolution. Despite these shortcomings, SIMS is still currently the most sensitive single-cell metabolic analysis method.
Using these methods, a significant number of metabolites have been identified in SCM studies. Among these metabolites are amino acids, phospholipids, tricarboxylic acid (TCA) cycle intermediates, nucleotides, fatty acids, and energy metabolites such as ATP [17]. Most of the studies have only identified metabolites present at high concentrations, such as the study by Bergman et al. in which the authors reported detection of several amino acids, including valine, histidine, proline, leucine/isoleucine, lysine, aspartic acid, glutamine, and glutamic acid at high levels in single human cells [29]. However, some studies have reported hundreds of different metabolites within a single cell besides those mentioned above. For instance, Sun et al. reported identifying over 550 metabolites including bilirubin, dehydrorabelomycin, and several avermectins in plant cells; however, they did not provide detailed information on the concentrations of the metabolites detects [30]. In another study by Pan et al., the authors detected many sphingomyelins, phosphatidylcholines, and phospholipids such as phosphoethanolamines at high concentrations in cervical cancer HeLa cells [31]. However, there are also phospholipids that are detected at low intensities, such as the phosphatidylglycerol, PG (34:1) [31]. Using their dicationic reagents, they were able to detect PG (34:1) at a higher intensity as well as to detect metabolites such as adenosine monophosphate (AMP), phosphatidic acids, and folic acid [17,31]. Of note, the concentration of any given metabolite could vary between different samples.
2. Impactful Findings Generated Using Single-cell Metabolomics in Divergent Fields
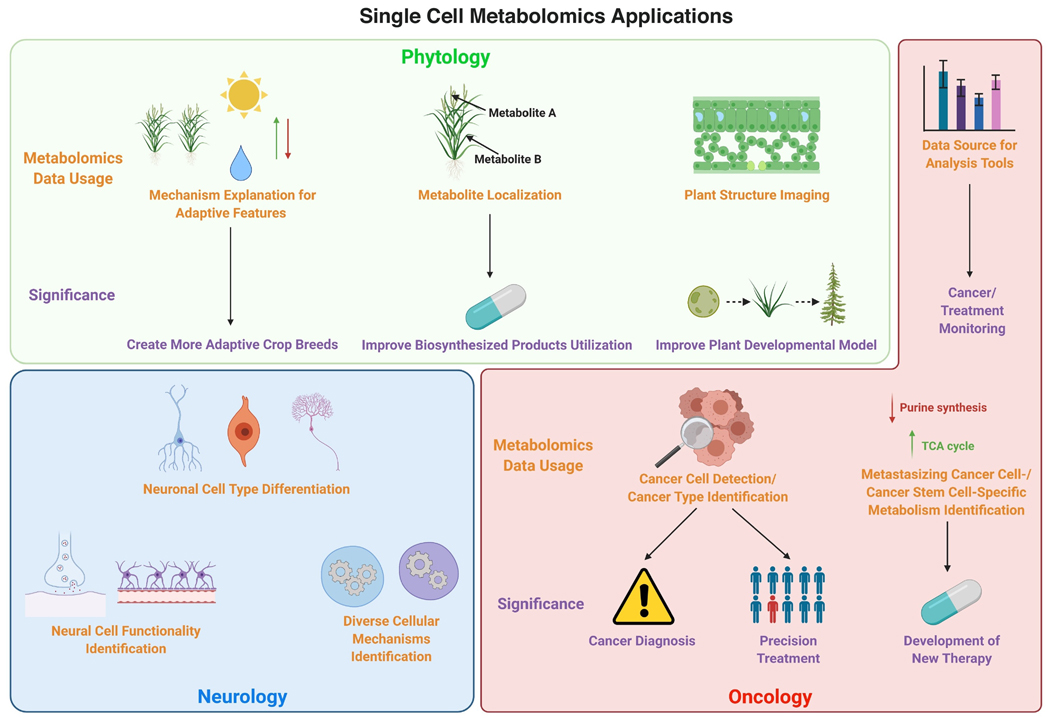
As a growing and powerful analytical tool, SCM has been applied to a wide range of fields. For example, live SCM using laser-desorption/ionization MS (LDI-MS) enables researchers to assess the physiological status of different microalgae widely found to form multi-species communities in both freshwater and marine systems [32]. This provides an opportunity to understand the diversity of species within a homogenous environment and explain how certain species can thrive despite selective feeding requirements and attack by pathogens while others die out. Metabolic analysis of cells from live frog embryos using CE-ESI-MS offers insight into the formation of single-cell metabolic heterogeneity at the early stages of vertebrate embryogenesis for the first time, refining vertebrate’s developmental biology [33]. Until now, SCM has shown its power and potential for analyzing diverse biological samples, supported by already available findings and data-verified hypotheses [34]. Its ability to analyze heterogeneous cells individually has provided it with the potential to be utilized in most fields of biology. This section will pay close attention to recent applications of SCM in phytology, neurology, and oncology (Figure 2).
Fig 2.
Single Cell Metabolomics’ Applications in Different Fields. Due to its exceptional power to carry out high resolution metabolic analysis, single cell metabolomics has great potential and applications in a variety of fields, including phytology, neurology, and oncology. Created with BioRender.com
2.1. Single-cell Metabolomics in Plant Research
SCM is widely used in phytology and has generated important findings. It provides the opportunity for the identification and metabolic profiling of cell types and organs in plants. Comparative metabolomics at the single-cell level reveals the change in epidermal bladder cells (EBCs) of halophyte M. crystallinum upon salt stress, explaining one of its adaptive features [35]. Similarly, another study uses microcapillary-assisted metabolite extraction followed by gas chromatography-MS (GC-MS) and ultrahigh-pressure liquid chromatography-MS/MS to reveal the developmental relationship between two types of trichomes of cannabis. The result supports the cannabis flower’s unique psychoactive and medicinal usage from a metabolic perspective [36]. Therefore, the ability of single-cell metabolomics to perform precise metabolic analysis helps offer explanations and mechanisms of unique plant functions and favorable features, which could greatly benefit the development of applications based on those functions and features, such as creating more adaptive crop breeds or more effective extraction of desirable biosynthesized compounds.
Besides cell and organ metabolic profiling, SCM is also a powerful tool in spatial metabolic analysis. Recently, SCM has helped researchers reveal the localization of terpenoid indole alkaloids (TIAs) and their precursors within different types of cells of C. roseus, a medicinal plant well-known for producing antitumor drugs. This study by Yamamoto et al. shows that although most TIA precursors are found in epidermal cells, major TIAs are found in idioblast cells [37]. Another study from the same authors also utilizes single-cell MS and imaging MS to show that many TIA intermediates, such as catharanthine and serpentine, are found in idioblast and laticifer cells [38]. These findings have revealed the complex localization of intracellular TIAs and two novel places for TIA biosynthesis, which may improve antitumor drug extraction efficiency. Similar spatial analyses have also been done on plant organs and whole plants. For example, Hölscher et al. used LDI-MSI to show the localization of secondary metabolites in A. thaliana and Hypericum species at the single-cell level [39]. Their methods enabled organ- specific distribution of secondary metabolites in A.thaliana to be imaged at single-cell resolution. A study by Li et al. reports similar spatial metabolic analysis at the organ level in the rhizome of G. glabra using atmospheric pressure MALDI tandem MS imaging. The authors were able to detect and visualize the spatial distribution of important tissue-specific metabolites, including free flavonoids and saponins, at the cellular level [40]. The success of these studies highlights the importance of the development of single-cell metabolic analysis techniques and the ability they provide to carry out plant metabolic research on different levels at single-cell resolution. As more precise plant structure imaging and metabolic change monitoring become available, such monitoring may serve as a direct approach to assess the effect of experimental treatments on plants in a wide range of studies.
All in all, considering the great diversity of species, tissues, and cell types in phytology, SCM has been shown to be a powerful tool decoding this heterogeneity in the trend of pursuing higher resolution in plant research. The advances of SCM in plant research could further benefit the assessment of experimental treatments, the understanding of favorable plant features, the construction of plant developmental biology models, and the medical use of plant-derived products.
2.2. Single-cell Metabolomics Helps Decipher Neural Cell Heterogeneity
Another field where SCM is widely applied is neurology. Neurons are often larger than other normal cells and have very detailed subtypes, and changes in neurological systems induced by disease or signals are often reflected in single cells [41]. Therefore, SCM is critical and valuable in metabolic profiling and heterogeneity assessment in neurological research.
A study by Nemes et al. used single-cell metabolic data of B1 and B2 buccal neurons from A. californica to show that these two cell types regulate their metabolome differently in response to changes in the external environment [42]. This study reveals the difference in cell response mechanisms in different neurons. It also shows that environmental factors play an important role in the formation of cell heterogeneity. The second finding is especially crucial as differences caused by culture processes may lead to false conclusions in many studies and cause difficulty replicating results. In addition, SCM promotes the examination of cell heterogeneity and cell type differentiation in neurological systems. In their study, Aerts et al. successfully showed the connection between physiological activity and neurochemical states of neurons and astrocytes using single-cell cytoplasm metabolomics [42].
Further, they showed the occurrence of significant cell-to-cell differences within the brain [43]. Similarly, differences among divergent neuronal cell types can also be assessed, as reported by Do et al., where they use single-cell profiling of lipids and other metabolites to effectively classify populations and subpopulations of neuronal cells [44]. These findings strongly support single-cell the power of metabolomics in deciphering neuronal cell heterogeneity, which provides more accurate differentiation between neural cell types and identifies specific functionalities and diverse internal cell mechanisms of different cell types in the neurological system.
2.3. Single-cell Metabolomics is Widely Used in Cancer Cell Identification
As a powerful technology capable of detecting and quantifying thousands of metabolites in various biological samples, metabolomics has already helped researchers further understand cancer metabolism and the key features of cancer cells. However, even within a single tumor, tumor cells could have very different morphologies and phenotypes, known as cancer heterogeneity [45]. Therefore, SCM is especially useful in oncology studies and may benefit multiple facets of research, including drug assessment, disease diagnosis, and precision drug design. Previous genomic analysis targeting metabolic genes at the single-cell level has revealed that individual malignant cells have elevated metabolic activity and variations that were not observed in bulk tumor studies [46].
Circulating tumor cells (CTCs) are cells that detach from the original tumor and enter the bloodstream. They convey rich information about their original tumors and play important roles in metastasis [47]. For example, SCM helps obtain the metabolic profile of CTCs in neuroblastoma patients [48]. Another recent SCM study on circulating melanoma cells revealed that purine synthesis is downregulated in these cells compared to tumor cells, potentially helping these cells maintain the level of NADPH required to mediate oxidative stress during metastasis [50]. This can potentially offer insights into preventing and treating metastatic cancer. Therefore, SCM shows great potential in not only disease detection and cancer prognosis but also helps provide new information important for the development of new treatments. Besides, SCM also plays an important role in cell identification. Zhang et al. recently reported successful metabolite profiling and differentiation between human astrocyte cells and glioblastoma cells at the single-cell level using their drop extraction and pulsed direct current ESI-MS (Pico-ESI-MS) method [51]. Another study also reported successful differentiation between breast cancer subtypes using single-cell metabolic data [52]. As a well-known feature of cancer, heterogeneity among different cancers and cancer subtypes has long posed a great challenge for effective treatment. Therefore, the potential of SCM in identifying cancer types/subtypes could significantly improve cancer treatment efficiency and promote the development of personalized treatment strategies. Similarly, Sun et al. reported differences between metabolic profiles of cancer stem cells (CSCs) and non-stem cancer cells after evaluating the metabolic characteristics of live colorectal CSCs at a single-cell resolution where the TCA cycle is upregulated [53]. This offers insights into the design of cancer treatments to combat CSCsr. Differentiating among cell subtypes using SCM sometimes requires pretreatment. A study by Fang et al. showed that mannose as a stimulant could effectively improve metabolic discrimination of osteosarcoma cells from normal human osteoblasts at the single-cell level [54]. These findings show that SCM has great value in decoding cancer cell heterogeneity.
Besides distinguishing cell types, SCM also leads to discoveries that could shape future treatment. Using mass cytometry, Harmann et al. profiled the metabolic regulome of human cytotoxic T cells and identified tissue-specific metabolically repressed cytotoxic T cells in human colorectal carcinoma [55]. This finding revealed the spatial arrangement of human tissue metabolic activity and immune repression occurring at the tumor-immune boundary. Also, using mass cytometry, Levine et al. identified a distinct metabolic state of T cells during an early immune response. Both studies demonstrate the ability of mass cytometry in SCM and how metabolic manipulation could help regulate the immune system and combat adverse immune responses [56].
SCM is also often used in combination with other cutting-edge analytical methods. For example, Liu et al. reported a combined use of SCM with machine learning for early monitoring of drug resistance induced by chemotherapy from a metabolomics point of view in cancer cells [57]. Therefore, metabolic data obtained from SCM could serve as an important data source for ever more sophisticated analysis to combat cancer.
All in all, SCM is a powerful tool to detect, identify and differentiate specific cancer cell types, which could significantly benefit cancer diagnosis and lead to precise and efficient treatment in response to different cancer cell types, as well as improve future research aimad at curing specific cancer types.
Conclusion
Obtaining metabolic data at the single-cell level poses many challenges, including low absolute metabolite concentrations for some important metabolites, the inability to amplify metabolites unlike RNA or DNA, fast conversion of metabolites, and a large difference in metabolite concentrations per cell [9]. Yet, efficient techniques have been developed to carry out sensitive, high throughput metabolomics analysis at the single-cell level. With further improvements and innovations in such techniques, SCM has enormous potential for carrying out single-cell resolution research in multiple fields, supported by already abundant findings. With its exceptional power to identify and differentiate single cells with varying metabolic profiles, SCM has already helped solve problems previously caused by the inability to separately analyze cells from the same organ or cluster. It enables us to understand plant features, decode characteristics of different neural cells, and combat cancer cell heterogeneity, and more. To pursue more detailed and specific profiling of the world around us, SCM is undoubtedly one of the most powerful tools we currently have available.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants R01-CA193895, R01-CA112314, 1S10OD025226-01, and UL1 TR001079 (to A.L). Special thanks to Dr. Arthur J. L. Cooper for his helpful editing.
Footnotes
Conflict of interest statement
The authors have declared no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Shenghao Guo, Department of Biomedical Engineering, Johns Hopkins University Whiting School of Engineering, Baltimore, MD, USA.
Cissy Zhang, Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD, USA; Department of Biology, Johns Hopkins University Krieger School of Arts & Sciences, Baltimore, MD, USA.
Anne Le, Department of Pathology and Oncology, Johns Hopkins University School of Medicine; Department of Chemical and Biomolecular Engineering, Johns Hopkins University Whiting School of Engineering, Baltimore, MD, USA.
References
- [1].Zenobi R: Single-cell metabolomics: analytical and biological perspectives. Science 2013, 342:1243259. [DOI] [PubMed] [Google Scholar]
- [2].Nguyen T, Kirsch BJ, Asaka R, Nabi K, Quinones A, Tan J, Antonio MJ, Camelo F, Li T, Nguyen S, et al. : Uncovering the Role of N-Acetyl-Aspartyl-Glutamate as a Glutamate Reservoir in Cancer. Cell Rep 2019, 27:491–501 e496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Luan H, Wang X, Cai Z: Mass spectrometry-based metabolomics: Targeting the crosstalk between gut microbiota and brain in neurodegenerative disorders. Mass Spectrom Rev 2019, 38:22–33. [DOI] [PubMed] [Google Scholar]
- [4].Paul K, Sorrentino M, Lucini L, Rouphael Y, Cardarelli M, Bonini P, Reynaud H, Canaguier R, Trtilek M, Panzarova K, et al. : Understanding the Biostimulant Action of Vegetal-Derived Protein Hydrolysates by High-Throughput Plant Phenotyping and Metabolomics: A Case Study on Tomato. Front Plant Sci 2019, 10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Pan Z, Raftery D: Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem 2007, 387:525–527. [DOI] [PubMed] [Google Scholar]
- [6].Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, et al. : Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP: Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A 2008, 105:13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang D, Bodovitz S: Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol 2010, 28:281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rubakhin SS, Lanni EJ, Sweedler JV: Progress toward single cell metabolomics. Curr Opin Biotechnol 2013, 24:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hansen RL, Lee YJ: High-Spatial Resolution Mass Spectrometry Imaging: Toward Single Cell Metabolomics in Plant Tissues. Chem Rec 2018, 18:65–77. [DOI] [PubMed] [Google Scholar]
- [11].Fujii T, Matsuda S, Tejedor ML, Esaki T, Sakane I, Mizuno H, Tsuyama N, Masujima T: Direct metabolomics for plant cells by live single-cell mass spectrometry. Nat Protoc 2015, 10:1445–1456. [DOI] [PubMed] [Google Scholar]
- [12].Rubakhin SS, Romanova EV, Nemes P, Sweedler JV: Profiling metabolites and peptides in single cells. Nat Methods 2011, 8:S20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Amantonico A, Urban PL, Zenobi R: Analytical techniques for single-cell metabolomics: state of the art and trends. Anal Bioanal Chem 2010, 398:2493–2504. [DOI] [PubMed] [Google Scholar]
- [14].Gong X, Zhao Y, Cai S, Fu S, Yang C, Zhang S, Zhang X: Single Cell Analysis with Probe ESI-Mass Spectrometry: Detection of Metabolites at Cellular and Subcellular Levels. Analytical Chemistry 2014, 86:3809–3816. [DOI] [PubMed] [Google Scholar]
- [15].Masuda K, Abouleila Y, Ali A, Yanagida T, Masujima T: Live Single-Cell Mass Spectrometry (LSC-MS) for Plant Metabolomics. Methods Mol Biol 2018, 1778:269–282. [DOI] [PubMed] [Google Scholar]
- [16].Mizuno H, Tsuyama N, Harada T, Masujima T: Live single-cell video-mass spectrometry for cellular and subcellular molecular detection and cell classification. J Mass Spectrom 2008, 43:1692–1700. [DOI] [PubMed] [Google Scholar]
- [17].Duncan KD, Fyrestam J, Lanekoff I: Advances in mass spectrometry based single-cell metabolomics. Analyst 2019, 144:782–793. [DOI] [PubMed] [Google Scholar]
- [18].Amantonico A, Urban PL, Fagerer SR, Balabin RM, Zenobi R: Single-cell MALDI-MS as an analytical tool for studying intrapopulation metabolic heterogeneity of unicellular organisms. Anal Chem 2010, 82:7394–7400. [DOI] [PubMed] [Google Scholar]
- [19].Dietmair S, Timmins NE, Gray PP, Nielsen LK, Kromer JO: Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal Biochem 2010, 404:155–164. [DOI] [PubMed] [Google Scholar]
- [20].Sellick CA, Hansen R, Maqsood AR, Dunn WB, Stephens GM, Goodacre R, Dickson AJ: Effective quenching processes for physiologically valid metabolite profiling of suspension cultured Mammalian cells. Anal Chem 2009, 81:174–183. [DOI] [PubMed] [Google Scholar]
- [21].Chen MM, Li AL, Sun MC, Feng Z, Meng XC, Wang Y: Optimization of the quenching method for metabolomics analysis of Lactobacillus bulgaricus. J Zhejiang Univ Sci B 2014, 15:333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ali A, Abouleila Y, Shimizu Y, Hiyama E, Emara S, Mashaghi A, Hankemeier T: Single-cell metabolomics by mass spectrometry: Advances, challenges, and future applications. TrAC Trends in Analytical Chemistry 2019, 120:115436. [Google Scholar]
- [23].Ong T-H, Kissick DJ, Jansson ET, Comi TJ, Romanova EV, Rubakhin SS, Sweedler JV: Classification of Large Cellular Populations and Discovery of Rare Cells Using Single Cell Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Analytical Chemistry 2015, 87:7036–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Amantonico A, Urban PL, Fagerer SR, Balabin RM, Zenobi R: Single-Cell MALDI-MS as an Analytical Tool for Studying Intrapopulation Metabolic Heterogeneity of Unicellular Organisms. Analytical Chemistry 2010, 82:7394–7400. [DOI] [PubMed] [Google Scholar]
- [25].Heinemann M, Zenobi R: Single cell metabolomics. Curr Opin Biotechnol 2011, 22:26–31. [DOI] [PubMed] [Google Scholar]
- [26].Lapainis T, Rubakhin SS, Sweedler JV: Capillary Electrophoresis with Electrospray Ionization Mass Spectrometric Detection for Single-Cell Metabolomics. Analytical Chemistry 2009, 81:5858–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Kawai T, Ota N, Okada K, Imasato A, Owa Y, Morita M, Tada M, Tanaka Y: Ultrasensitive Single Cell Metabolomics by Capillary Electrophoresis–Mass Spectrometry with a Thin-Walled Tapered Emitter and Large-Volume Dual Sample Preconcentration. Analytical Chemistry 2019, 91:10564–10572. ** The authors improved the resolution of capillary electrophoresis–mass spectrometry (CE-MS) using their self-designed ionization emitter and accomplished single-cell metabolic analysis on HeLa cells. This study demonstrates the constant advances made in single-cell metabolomics technologies.
- [28].Zhang XC, Wei ZW, Gong XY, Si XY, Zhao YY, Yang CD, Zhang SC, Zhang XR: Integrated Droplet-Based Microextraction with ESI-MS for Removal of Matrix Interference in Single-Cell Analysis. Sci Rep 2016, 6:24730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bergman HM, Lanekoff I: Profiling and quantifying endogenous molecules in single cells using nano-DESI MS. Analyst 2017, 142:3639–3647. [DOI] [PubMed] [Google Scholar]
- [30].Sun M, Yang Z, Wawrik B: Metabolomic Fingerprints of Individual Algal Cells Using the Single-Probe Mass Spectrometry Technique. Front Plant Sci 2018, 9:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pan N, Rao W, Standke SJ, Yang Z: Using Dicationic Ion-Pairing Compounds To Enhance the Single Cell Mass Spectrometry Analysis Using the Single-Probe: A Microscale Sampling and Ionization Device. Anal Chem 2016, 88:6812–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Baumeister TUH, Vallet M, Kaftan F, Svatos A, Pohnert G: Live Single-Cell Metabolomics With Matrix-Free Laser/Desorption Ionization Mass Spectrometry to Address Microalgal Physiology. Front Plant Sci 2019, 10:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Onjiko RM, Portero EP, Moody SA, Nemes P: In Situ Microprobe Single-Cell Capillary Electrophoresis Mass Spectrometry: Metabolic Reorganization in Single Differentiating Cells in the Live Vertebrate (Xenopus laevis) Embryo. Anal Chem 2017, 89:7069–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tian J, Fu G, Xu Z, Chen X, Sun J, Jin B: Urinary exfoliated tumor single-cell metabolomics technology for establishing a drug resistance monitoring system for bladder cancer with intravesical chemotherapy. Med Hypotheses 2020, 143:110100. [DOI] [PubMed] [Google Scholar]
- [35].Barkla BJ, Vera-Estrella R: Single cell-type comparative metabolomics of epidermal bladder cells from the halophyte Mesembryanthemum crystallinum. Front Plant Sci 2015, 6:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Livingston SJ, Quilichini TD, Booth JK, Wong DCJ, Rensing KH, Laflamme-Yonkman J, Castellarin SD, Bohlmann J, Page JE, Samuels AL: Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J 2020, 101:37–56. [DOI] [PubMed] [Google Scholar]
- [37]. Yamamoto K, Takahashi K, Caputi L, Mizuno H, Rodriguez-Lopez CE, Iwasaki T, Ishizaki K, Fukaki H, Ohnishi M, Yamazaki M, et al. : The complexity of intercellular localisation of alkaloids revealed by single-cell metabolomics. New Phytol 2019, 224:848–859. ** The authors used single-cell metabolomics to identify where terpenoid indole alkaloids (TIAs) and their precursors are localized within epidermal, idioblast, and laticifer cells of C. roseus. This study demonstrates the potential of single-cell metabolomics in plant metabolic research.
- [38].Yamamoto K, Takahashi K, Mizuno H, Anegawa A, Ishizaki K, Fukaki H, Ohnishi M, Yamazaki M, Masujima T, Mimura T: Cell-specific localization of alkaloids in Catharanthus roseus stem tissue measured with Imaging MS and Single-cell MS. Proc Natl Acad Sci U S A 2016, 113:3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hölscher D, Shroff R, Knop K, Gottschaldt M, Crecelius A, Schneider B, Heckel DG, Schubert US, Svatoš A: Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. The Plant Journal 2009, 60:907–918. [DOI] [PubMed] [Google Scholar]
- [40].Li B, Bhandari DR, Janfelt C, Römpp A, Spengler B: Natural products in Glycyrrhiza glabra (licorice) rhizome imaged at the cellular level by atmospheric pressure matrix-assisted laser desorption/ionization tandem mass spectrometry imaging. The Plant Journal 2014, 80:161–171. [DOI] [PubMed] [Google Scholar]
- [41].Qi M, Philip MC, Yang N, Sweedler JV: Single Cell Neurometabolomics. ACS Chem Neurosci 2018, 9:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV: Single-cell metabolomics: changes in the metabolome of freshly isolated and cultured neurons. ACS Chem Neurosci 2012, 3:782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Aerts JT, Louis KR, Crandall SR, Govindaiah G, Cox CL, Sweedler JV: Patch Clamp Electrophysiology and Capillary Electrophoresis–Mass Spectrometry Metabolomics for Single Cell Characterization. Analytical Chemistry 2014, 86:3203–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Do TD, Comi TJ, Dunham SJB, Rubakhin SS, Sweedler JV: Single Cell Profiling Using Ionic Liquid Matrix-Enhanced Secondary Ion Mass Spectrometry for Neuronal Cell Type Differentiation. Analytical Chemistry 2017, 89:3078–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tellez-Gabriel M, Ory B, Lamoureux F, Heymann MF, Heymann D: Tumour Heterogeneity: The Key Advantages of Single-Cell Analysis. Int J Mol Sci 2016, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Xiao Z, Dai Z, Locasale JW: Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun 2019, 10:3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Plaks V, Koopman CD, Werb Z: Cancer. Circulating tumor cells. Science 2013, 341:1186–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hiyama E, Ali A, Amer S, Harada T, Shimamoto K, Furushima R, Abouleila Y, Emara S, Masujima T: Direct Lipido-Metabolomics of Single Floating Cells for Analysis of Circulating Tumor Cells by Live Single-cell Mass Spectrometry. Anal Sci 2015, 31:1215–1217. [DOI] [PubMed] [Google Scholar]
- [49].Del Ben F, Turetta M, Celetti G, Piruska A, Bulfoni M, Cesselli D, Huck WT, Scoles G: A Method for Detecting Circulating Tumor Cells Based on the Measurement of Single-Cell Metabolism in Droplet-Based Microfluidics. Angew Chem Int Ed Engl 2016, 55:8581–8584. [DOI] [PubMed] [Google Scholar]
- [50].DeVilbiss AW, Zhao Z, Martin-Sandoval MS, Ubellacker JM, Tasdogan A, Agathocleous M, Mathews TP, Morrison SJ: Metabolomic profiling of rare cell populations isolated by flow cytometry from tissues. Elife 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang XC, Zang Q, Zhao H, Ma X, Pan X, Feng J, Zhang S, Zhang R, Abliz Z, Zhang X: Combination of Droplet Extraction and Pico-ESI-MS Allows the Identification of Metabolites from Single Cancer Cells. Anal Chem 2018, 90:9897–9903. [DOI] [PubMed] [Google Scholar]
- [52]. Wang R, Zhao H, Zhang X, Zhao X, Song Z, Ouyang J: Metabolic Discrimination of Breast Cancer Subtypes at the Single-Cell Level by Multiple Microextraction Coupled with Mass Spectrometry. Analytical Chemistry 2019, 91:3667–3674. *** The authors used single-cell metabolomics to differentiate the subtypes of breast cancer. This study demonstrates how single-cell metabolomics can be used in cancer research to help researchers gather information on cancer heterogeneity and improve current cancer treatment strategies.
- [53].Sun M, Yang Z: Metabolomic Studies of Live Single Cancer Stem Cells Using Mass Spectrometry. Analytical Chemistry 2019, 91:2384–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Fang Z, Wang R, Zhao H, Yao H, Ouyang J, Zhang X: Mannose Promotes Metabolic Discrimination of Osteosarcoma Cells at Single-Cell Level by Mass Spectrometry. Analytical Chemistry 2020, 92:2690–2696. * The authors used single-cell metabolomics and the stimulant mannose to help differentiate osteosarcoma cells from normal human osteoblasts. This study demonstrates how researchers can use pre-treatment as a way to improve single-cell metabolomics.
- [55].Hartmann FJ, Mrdjen D, McCaffrey E, Glass DR, Greenwald NF, Bharadwaj A, Khair Z, Verberk SGS, Baranski A, Baskar R, et al. : Single-cell metabolic profiling of human cytotoxic T cells. Nat Biotechnol 2021, 39:186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Levine LS, Hiam-Galvez KJ, Marquez DM, Tenvooren I, Madden MZ, Contreras DC, Dahunsi DO, Irish JM, Oluwole OO, Rathmell JC, et al. : Single-cell analysis by mass cytometry reveals metabolic states of early-activated CD8(+) T cells during the primary immune response. Immunity 2021, 54:829–844 e825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu R, Sun M, Zhang G, Lan Y, Yang Z: Towards early monitoring of chemotherapy-induced drug resistance based on single cell metabolomics: Combining single-probe mass spectrometry with machine learning. Anal Chim Acta 2019, 1092:42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]