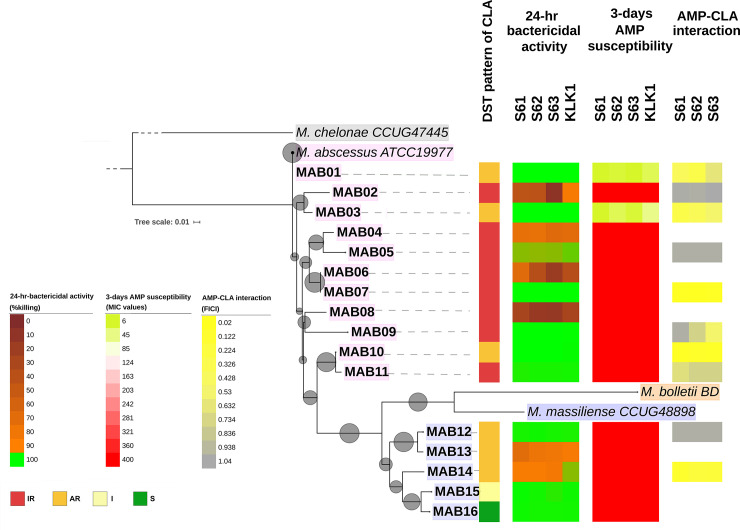
Fig 3. The phylogeny of M. abscessus and clarithromycin/ AMPs susceptibility patterns.
A whole genome-based tree of 1,000 bootstraps from 3,180 SNPs is shown. The sequences of reference strains M. chelonae CCUG47445, M. abscessus subsp. abscessus ATCC19977, M. abscessus subsp. bolletii BD, and M. abscessus subsp. massiliense CCUG48898 were included without phenotypic results. Ten clinical isolates as representatives from the phylogenetic tree including inducible and acquired resistance of two Mab subspecies were selected for the AMP-clarithromycin interaction assay. AR, acquired resistance; CLA, clarithromycin; DST, drug susceptibility testing; FICI, fractional inhibitory concentration index; I, intermediate; IR, inducible resistance; MIC, minimum inhibitory concentration; S, susceptible.