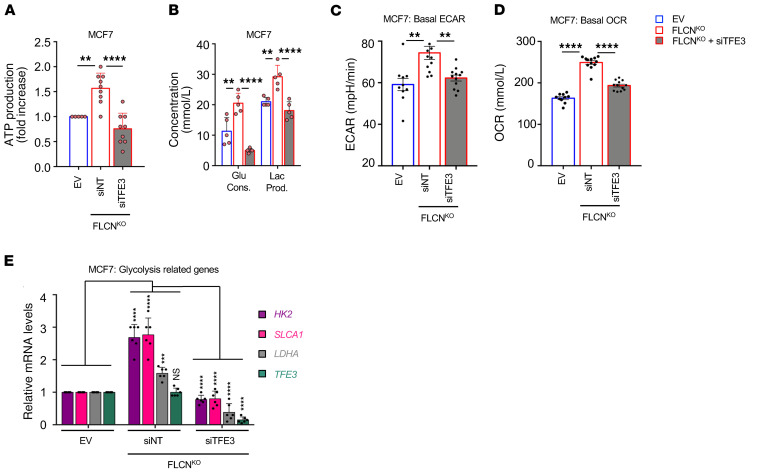
Figure 3. Loss of FLCN in MCF7 cells enhances cellular metabolism in a TFE3-dependent manner.
(A) Fold change in ATP levels in empty vector (EV) and FLCN-knockout (FLCNKO) MCF7 cells transfected with nontargeting (NT) siRNA control or siRNA targeting TFE3, after 48 hours of transfection as measured by CellTiter-Glo Luminescent Cell Viability Assay. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. **P < 0.01; ****P < 0.0001. (B) Glucose consumption and lactate production levels in the cellular media were measured using a NOVA Bioanalysis flux analyzer in EV and FLCNKO MCF7 cells transfected with nontargeting (NT) control siRNA or siRNA targeting TFE3 after 48 hours of transfection. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. **P < 0.01; ****P < 0.0001. (C and D) Basal extracellular acidification rate (ECAR) (C) and oxygen consumption rate (OCR) (D) in EV and FLCNKO MCF7 cells transfected with NT control siRNA or siRNAs targeting TFE3, after 48 hours of transfection, measured by Seahorse Bioscience XF96 extracellular flux analyzer. Data represent the average ± SEM of at least n = 3 independent experiments, each performed in triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. **P < 0.01; ****P < 0.0001. (E) Relative mRNA levels of TFE3 and glycolysis-related genes measured by RT-qPCR in EV and FLCNKO MCF7 cells transfected with nontargeting (NT) control siRNA or siRNA targeting TFE3. Data represent the average ± SEM of n = 6 independent experiments, each performed in triplicate, where each point represents the average of the triplicate. Statistical significance was determined using 2-way ANOVA with Bonferroni’s multiple-comparison correction. ***P < 0.001; ****P < 0.0001. NS, not significant.