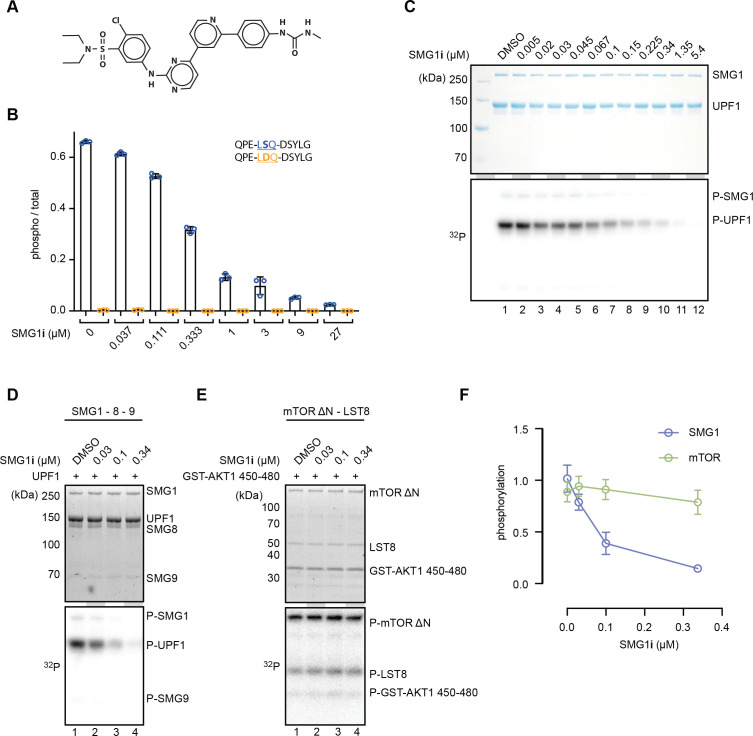
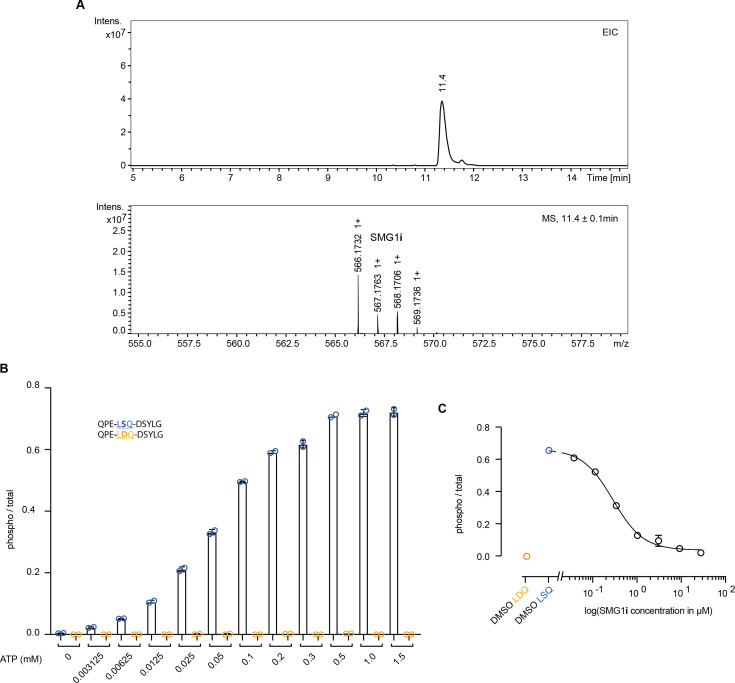
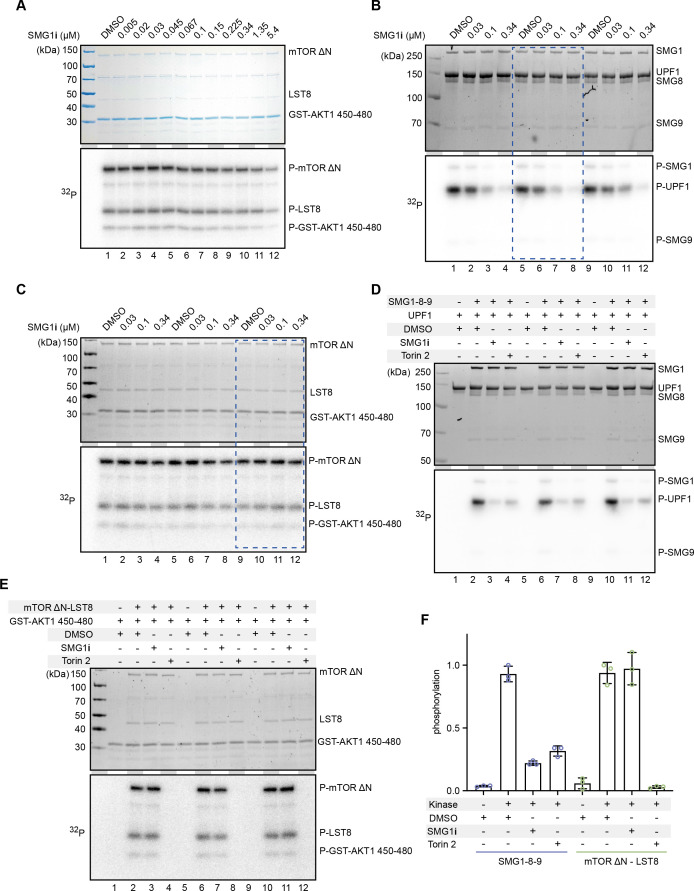
Figure 1. SMG1i specifically inhibits SMG1 kinase activity in vitro.
(A) Structure of the SMG1 inhibitor (SMG1i). (B) Titration of SMG1i using a mass spectrometry-based phosphorylation assay with 500 nM SMG1-8-9 and the indicated UPF1-derived peptides as substrates. (C) Titration of SMG1i using a radioactivity-based phosphorylation assay with 100 nM SMG1-8-9 and full-length UPF1 as a substrate. The Coomassie-stained gel is shown on top, and the corresponding radioactive signal is below. (D, E) Titration of SMG1i using a radioactivity-based phosphorylation assay with either SMG1-8-9 and full-length UPF1 (D) or mTORΔN-LST8 and GST-AKT1 (E) as a substrate. Stain-free gels are shown on top and the corresponding radioactive signal at the bottom (Ladner et al., 2004). (F) Quantification of normalized UPF1 or mTOR (auto-)phosphorylation in the presence of increasing amounts of SMG1i for both SMG1 and mTOR. For each data point, the mean is shown with standard deviations of the three replicates indicated.