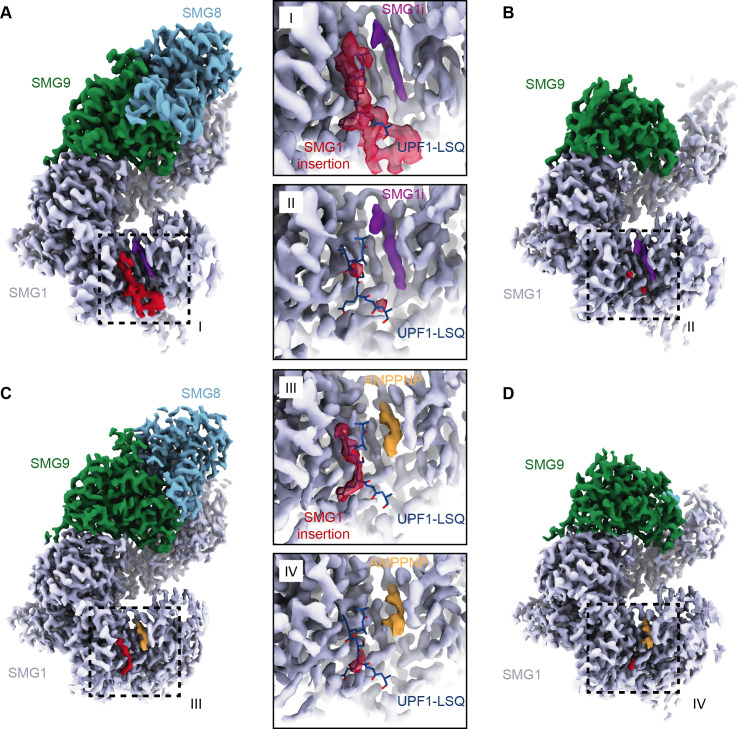
Figure 3. Structures of SMG1-8-9 and SMG1-9 complexes reveal that the SMG1 insertion domain can block the substrate-binding path in the presence of SMG8.
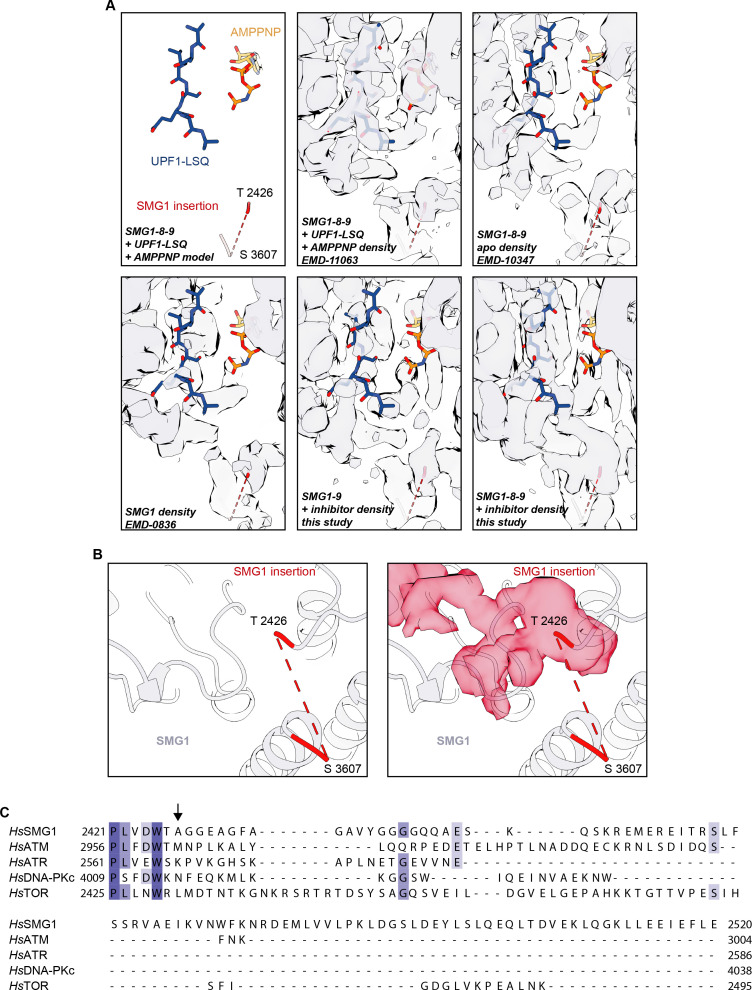
(A) Cryo-EM density of SMG1-8-9 bound to SMG1i. Density for the inhibitor is in magenta, the N-terminus of the SMG1 insertion is in red, and all other parts as indicated. (B) Cryo-EM density of SMG1-9 complex bound to the SMG1 inhibitor. Everything else as in (A). (C) Cryo-EM density of SMG1-8-9 bound to AMPPNP. Density for AMPPNP is in orange and extra density attributed to the SMG1 insertion is in red. (D) As in (C), but for the SMG1-9 complex bound to AMPPNP. Insets I–IV show close-ups of the indicated kinase active site densities superimposed with the model for the UPF1-LSQ substrate shown as blue sticks (PDB identifier: 6Z3R). All maps segmented. cryo-EM, cryo-electron microscopy.