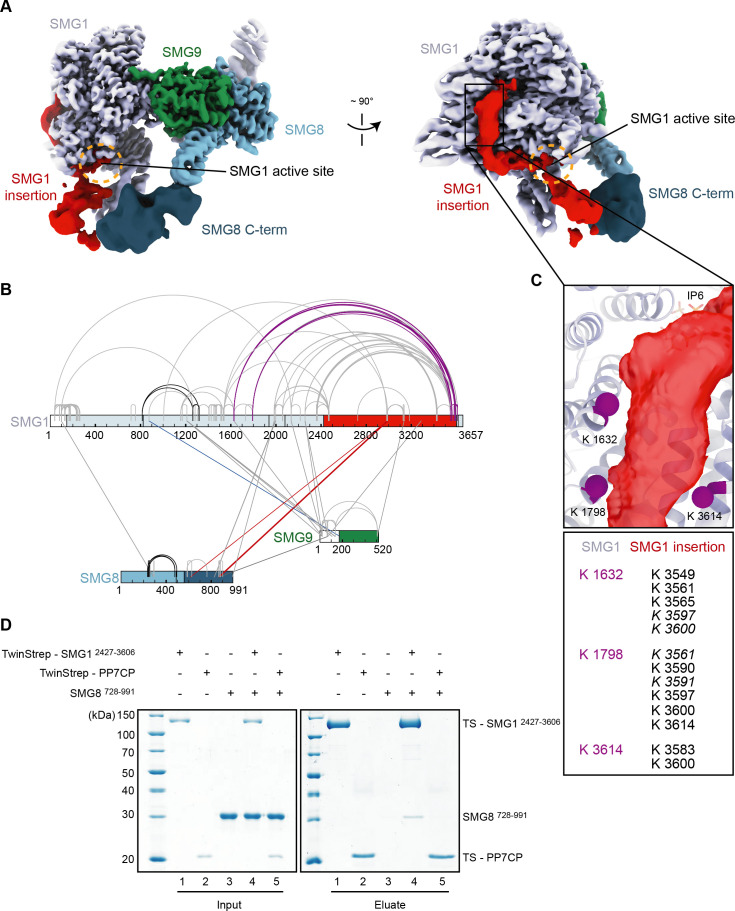
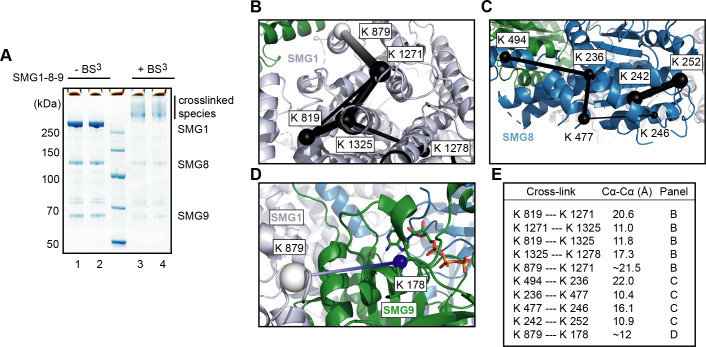
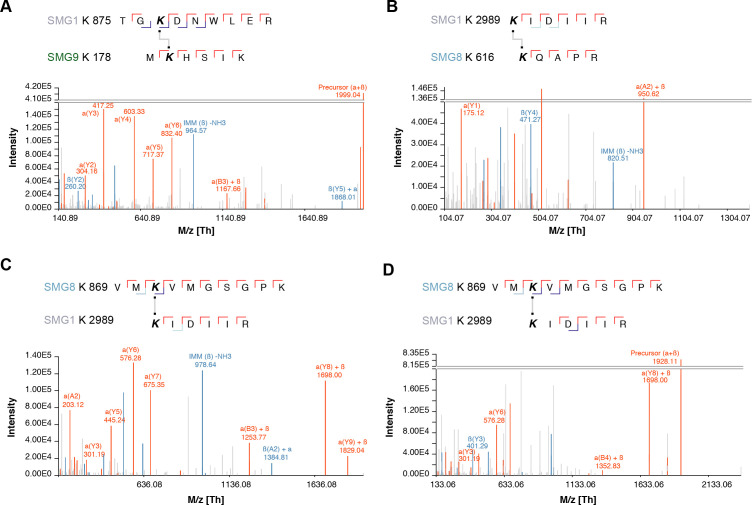
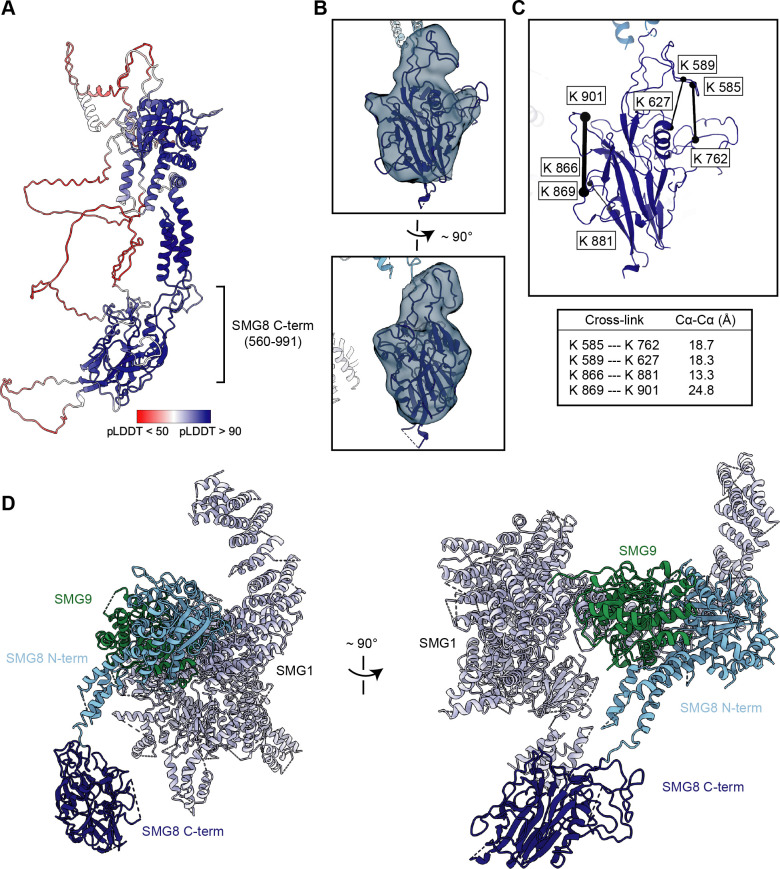
Figure 4. The SMG1 insertion domain can block overall access to the kinase active site.
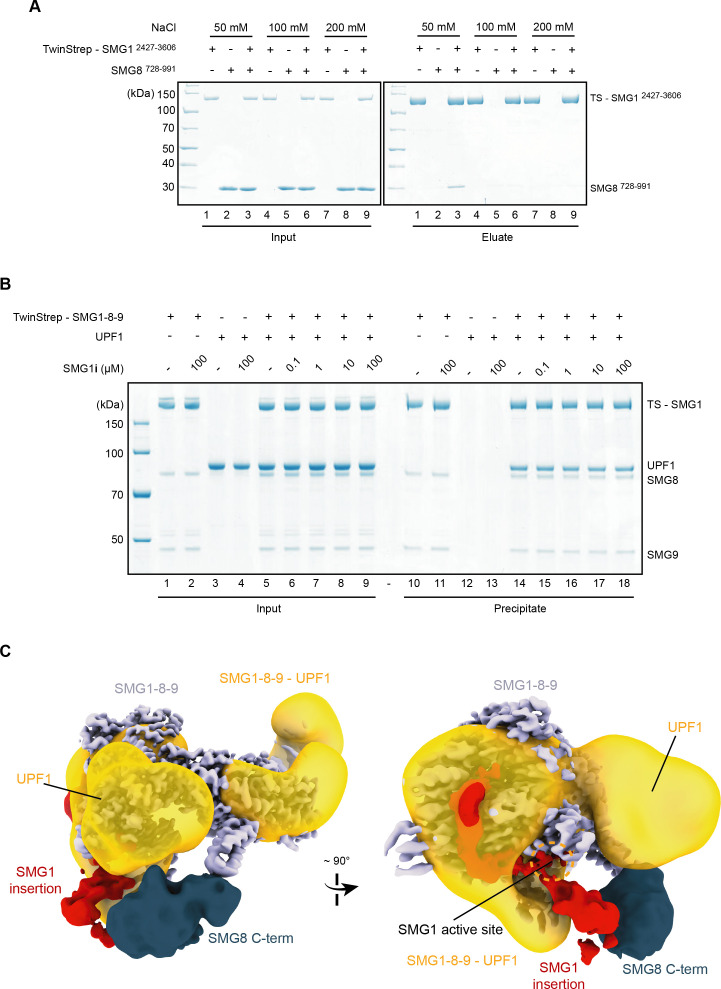
(A) Cryo-EM map after 3D variability analysis filtered by resolution and segmented. Two different views displaying extra density for SMG8 C-terminus (dark blue) and SMG1 insertion domain. The location of the SMG1 kinase active site is indicated by an orange circle. (B) Inter and intra cross-links of the SMG1-8-9 kinase complex. Proteins are colored as in (A). Intra-links shown in Figure 4—figure supplement 1B, C are in black, the inter-link shown in Figure 4—figure supplement 1D is in blue, and inter-links between SMG1 insertion and SMG1 C-terminus are in red. (C) Zoom-in highlighting positions of Lys residues (magenta spheres) crosslinking to the C-terminus of the SMG1 insertion domain. A table listing the cross-linked residues is shown below. Cross-links found in only one of two samples are in italics. (D) Coomassie-stained SDS-PAGE analysis of pull-down experiment showing an interaction between SMG1 insertion domain (SMG12427–3606) and SMG8 C-terminus (SMG8728–991). cryo-EM, cryo-electron microscopy.