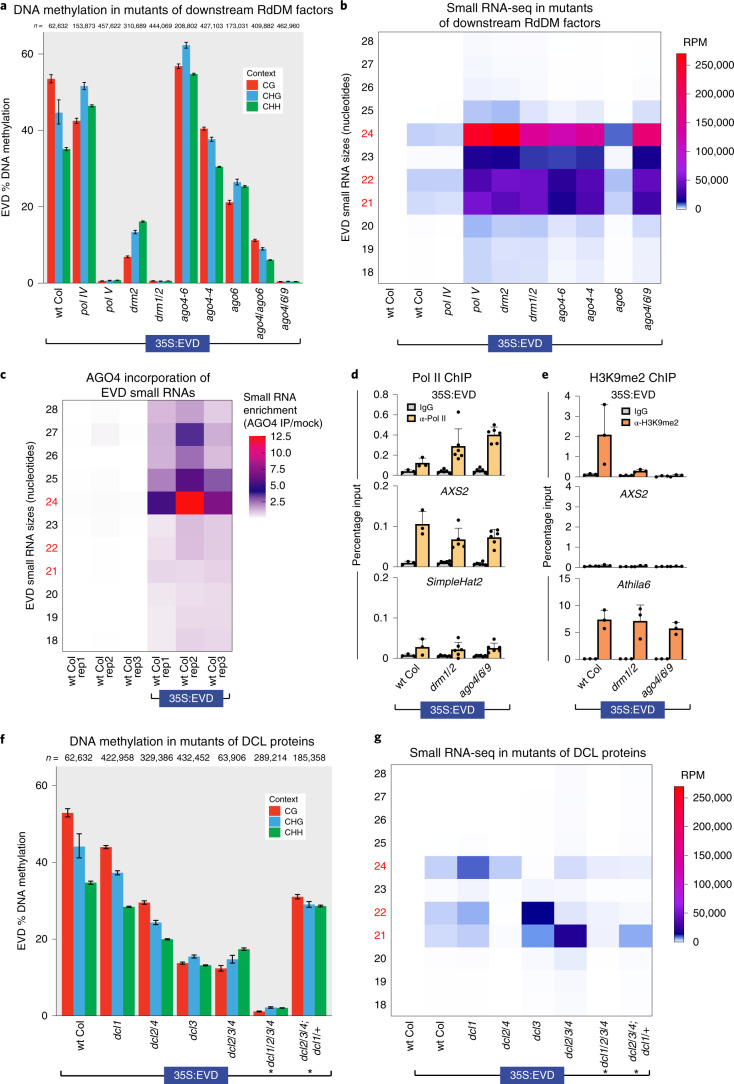
Fig. 2. Genetic requirements for the first round of RdDM.
a, BSAS of 35S:EVD in T1 plants lacking key RdDM factors demonstrates that the initiation of DNA methylation is dependent on Pol V, DRM1/2 and at least one member of the AGO4-clade of proteins. The data display is the same as Fig. 1a. The wt Col and pol V data are replicates 1 and 3, respectively, from Fig. 1a. b, Heatmap of small RNA sequencing for transgenic lines from a. The accumulation of differently sized siRNAs is shown on the y axis, with the key sizes 21, 22 and 24 nt in red. The siRNAs shown were mapped to the entire length of the EVD coding region, not just the region analysed in a with BSAS. c, Heatmap of EVD small RNA enrichment in AGO4. Enrichment is calculated as the ratio of small RNA accumulation (RPM) in AGO4-IP over mock-IP samples for each size class of small RNA. d, ChIP analysis of Pol II (Ser5P) in T1 transgenic plants from a shown as percentage input. Error bars represent the standard deviation of the mean. Three biological replicates were used for wt Col, and six biological replicates were used for the other two genotypes. SimpleHat2 is a transcriptionally silenced TE negative control, and AXS2 is a transcribed positive control gene. e, ChIP analysis of H3K9me2 levels in T1 transgenic plants shown as percentage input. Three biological replicates were used for each genotype. Athila6 is a silenced TE with known accumulation of H3K9me2. Error bars represent the standard deviation of the mean of three biological replicates. f, BSAS of 35S:EVD in T1 plants lacking various combinations of DCL family proteins. The dcl1/2/3/4 quadruple mutant plants are siblings of the dcl2/3/4; dcl1/+ plants (siblings are denoted with an asterisk). The data display is the same as Fig. 1a. The wt Col data are replicate 1 from Fig. 1a. g, Heatmap of small RNA sequencing for the same transgenic lines as in f. The wt Col data are the same from b.