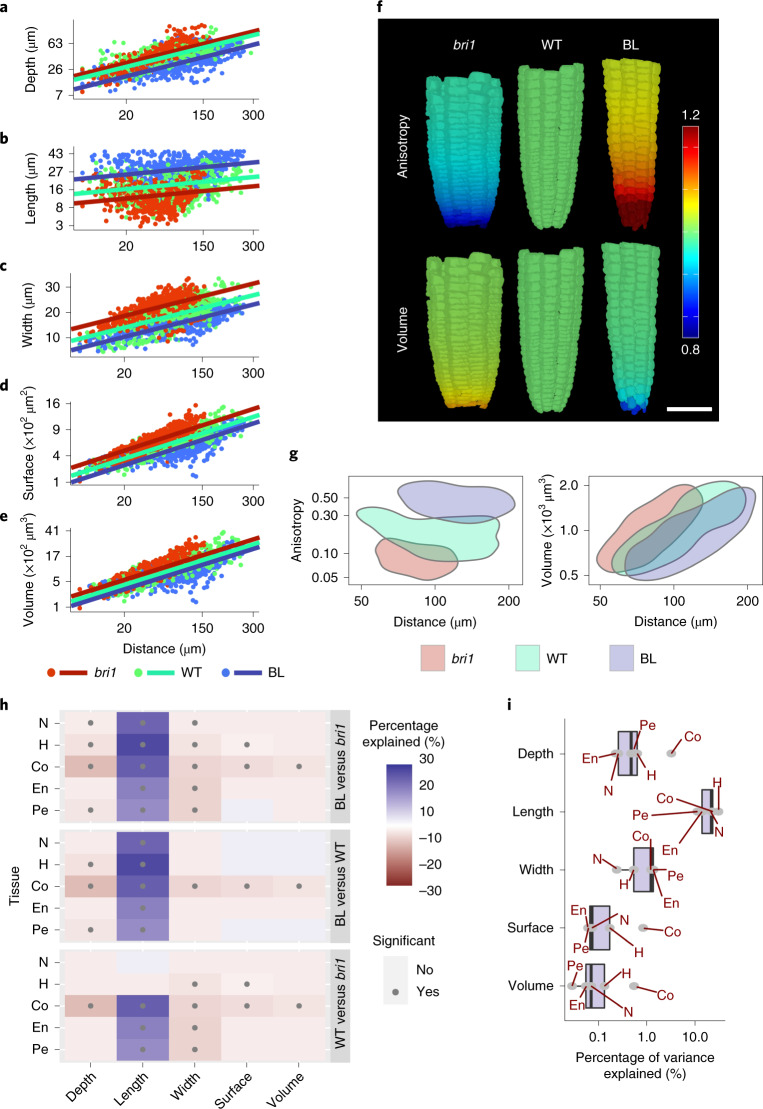
Fig. 2. BR signalling has a higher effect on cell shape and anisotropy than on cell volume across meristematic tissues.
a–e, Robust differences in geometric parameters of individual meristematic cells as captured by a mixed-model ANOVA (see the text for the details). Shown here are the effects of the distance from the QC (x axis, square-root-transformed) and BR signalling strength (differing among bri1, WT and BL-treated WT roots) on cell depth (log-transformed) (a), length (log-transformed) (b), width (c), surface area (square-root-transformed) (d) and volume (cube-root-transformed) (e) in the cortex. The axes are scaled according to the transformations. Corresponding analyses in other tissues are shown in Extended Data Fig. 2. See h for significant pairwise comparisons that result from this analysis. Note the opposite trend between roots with high and low BR signalling. Also note that all geometric parameters except length are higher in bri1. f,g, Comparison of anisotropy index (calculated as length2/(depth × width)) and volume between bri1, WT and BL-treated WT roots. In f, a display of the cortex tissue in representative segmented roots is shown, depicting relative differences in anisotropy index and volume (WT = 1) as a function of distance from the QC. In g, 2D kernel density plots of anisotropy index (left) and volume (cube-root-transformed, right) versus distance from the QC of WT, bri1 and BL-treated WT root cells are shown. Note that BL-treated cells have significantly higher anisotropy, while bri1 cells have significantly lower anisotropy. In contrast, the two groups of cells showed similar volume values. P < 0.05; mixed model. h, BR signalling has a higher effect on cell length, depth and width parameters than on cell volume across tissues. The heat map presents the percentage of variance explained by BR. Shown are all pairwise comparisons organized in three blocks (WT versus bri1, WT roots treated with BL versus WT and WT roots treated with BL versus bri1), for five geometric parameters in five root cell types and tissues. Blue indicates that the first treatment in the comparison has a higher value. Red indicates that the first treatment in the comparison has a lower value. The higher the opacity, the higher the percentage of variance explained. The dots indicate significance (adjusted P < 0.05; two-step adaptive correction; Methods). Note the significant, robust, opposing differences between bri1 and BL-treated samples in length, depth and width across tissues, while cell volume remained largely stable. i, Box plot summarizing the effect of treatment on geometric parameters in terms of percent variance explained by BR signalling in a given tissue (grey dots). The plot shows the interquartile range as the left and right boundaries of the box, the median as an internal vertical line, and the maximum and minimum values as whiskers. For all panels, n = 4 roots for WT, 4 roots for bri1 and 3 roots for BL. N, non-hair cells of the epidermis (n = 374 cells for WT, 376 cells for bri1 and 280 cells for BL); H, hair cells of the epidermis (n = 288 cells for WT, 322 cells for bri1 and 460 cells for BL); Co, cortex (n = 615 cells for WT, 532 cells for bri1 and 462 cells for BL); En, endodermis (n = 445 cells for WT, 389 cells for bri1 and 622 cells for BL); Pe, pericycle (n = 633 cells for WT, 749 cells for bri1 and 833 cells for BL).