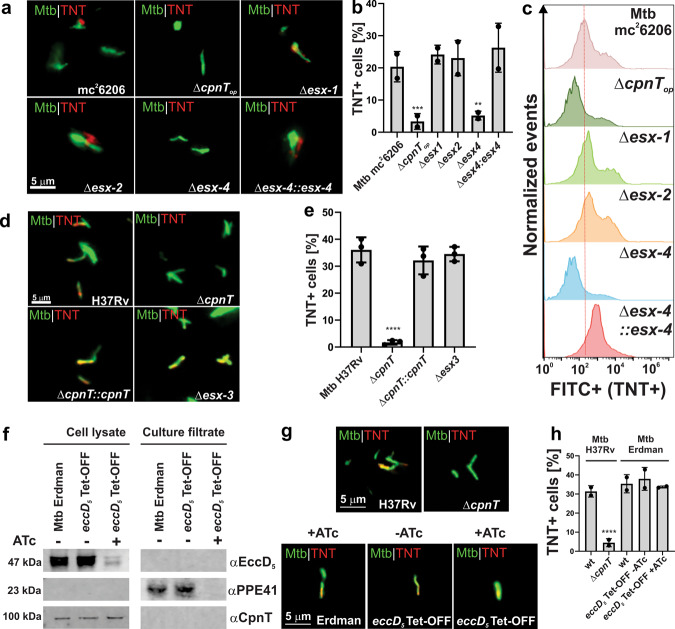
Fig. 2. The ESX-4 system is essential for CpnT export and surface accessibility in M. tuberculosis.
a Detection of surface-accessible CpnT in Mtb using fluorescence microscopy. The Mtb strains were labeled with DMN-trehalose (green) and stained with polyclonal anti-TNT and Alexa Fluor-594 secondary antibodies (red). b Quantification of TNT-positive Mtb cells from images shown in a. Mtb cells were scored as TNT-positive when a red signal was observed as compared with Mtb ΔcpnTop. c Surface accessibility of CpnT in Mtb by flow cytometry. The Mtb strains were stained with polyclonal anti-TNT and Alexa Fluor-488 secondary antibodies. The mean fluorescence of Mtb cells is displayed in histograms. d Surface-accessibility of CpnT of the Mtb H37Rv esx-3 deletion mutant and the control strains by fluorescence microscopy. The staining procedure was performed as in a. e Quantification of TNT-positive Mtb cells from images shown in d. Mtb cells were scored as TNT-positive when a red signal was observed as compared to Mtb ΔcpnT. f Depletion of a functional ESX-5 system in Mtb Erdman. The wt and eccD5 Tet-OFF mutant strains were grown in the presence/absence of anhydrotetracycline hydrochloride (ATc; 100 ng/ml). Proteins from the whole-cell lysates and culture filtrates were analyzed by immunoblotting to detect EccD5, PPE41, and CpnT with specific antibodies. g Surface-accessibility of CpnT in Mtb Erdman depleted of a functional Esx-5 system using fluorescence microscopy. The staining procedure was performed as in a. The wt and ΔcpnT strains in the H37Rv background were used as positive and negative controls, respectively. h TNT-positive Mtb cells from images shown in g were quantified as described in e. The Δesx-1 strain has a deletion of the RD1 region and lacks the panCD genes. Data are represented as mean ± SEM of at least two independent experiments (n ≥ 2) and representative images/blots are shown. Asterisks indicate significant differences (*p value ≤ 0.05, **p value ≤ 0.01, *** value ≤ 0.001, ****p value ≤ 0.0001, calculated using the one-way ANOVA with Dunnett’s correction) compared with the corresponding wt strain. Source data are provided in the Source Data file.