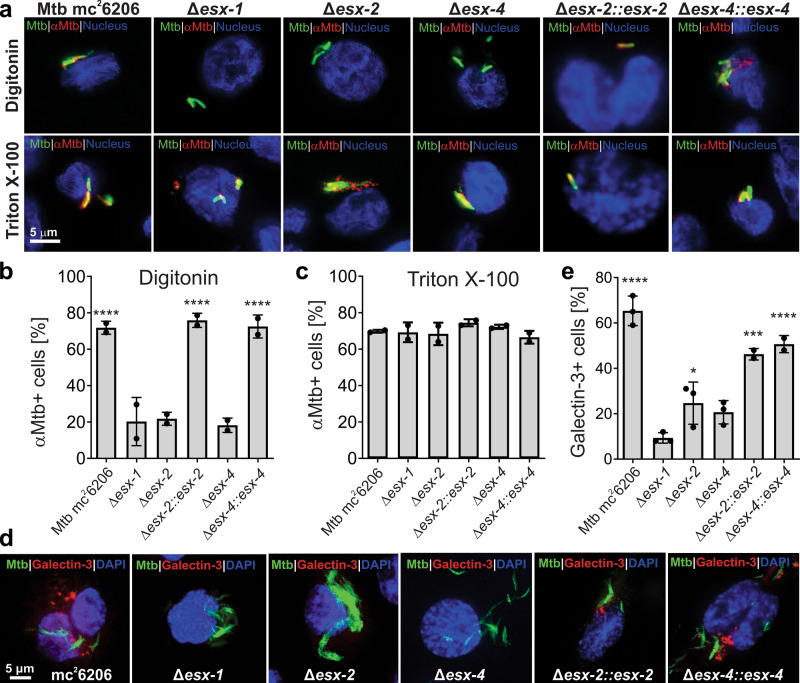
Fig. 5. The ESX-1, ESX-2, and ESX-4 systems are required for phagosomal rupture by M. tuberculosis.
a Detection of phagosomal rupture in infected macrophages by using Mtb-specific antibodies. The indicated Mtb strains were labeled with the metabolic dye DMN-Trehalose (green) and used to infect THP-1 macrophages at an MOI of 10:1. After 48 h of infection, the macrophages were permeabilized with digitonin to enable access of antibodies to the cytoplasm, or with Triton X-100 for access to intracellular compartments. Then, cells were stained with an anti-Mtb antibody (αMtb) and with an Alexa Fluor-594 secondary antibody (red). The macrophage nuclei were stained with DAPI. b, c Quantification of αMtb-positive macrophages after permeabilization with digitonin (b) and Triton X-100 (c) from images shown in a. Macrophages were scored as αMtb-positive when distinct red punctae were observed as compared to the Mtb esx-1 mutant treated with digitonin. d Detection of ruptured phagosomes in macrophages infected with Mtb. The indicated Mtb strains were labeled with the metabolic dye DMN-Trehalose (green) and used to infect THP-1 macrophages at an MOI of 10:1. After 48 h of infection, the macrophages were permeabilized with Triton X-100 and stained with an antibody against the phagosome rupture marker Galectin-3 and with an Alexa Fluor-594 secondary antibody (red). e. Quantification of Galectin-3-positive macrophages from images shown in d. Macrophages were scored as Galectin-3-positive when distinct red punctae were observed as compared to the Mtb esx-1 mutant. The Δesx-1 strain has a deletion of the RD1 region and lacks the panCD genes. Data are represented as mean ± SEM of at least two independent experiments (n ≥ 2) and representative images are shown. Asterisks indicate significant differences (*p value ≤ 0.05, **p value ≤ 0.01, ***p value ≤ 0.001, ****p value ≤ 0.0001, calculated using the one-way ANOVA with Dunnett’s correction) compared with the Mtb Δesx-1 strain. Source data are provided in the Source Data file.