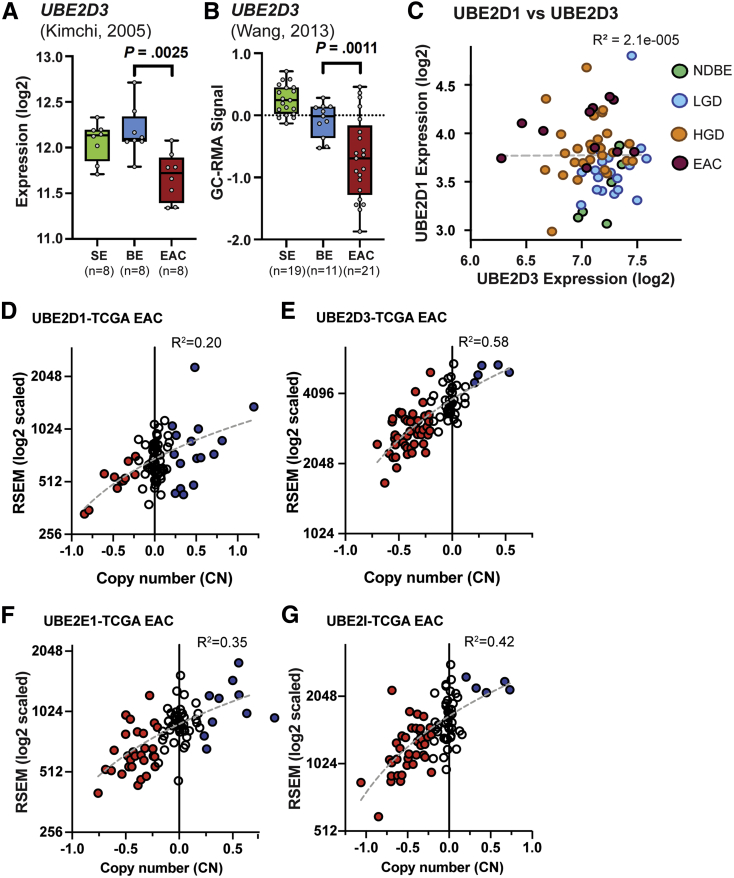
Figure 4.
Confirmation of UBE2D3 changes in other BE cohorts, and RNF128 to specific E2 copy number correlations in the ESCA TCGA cohort for esophageal cancer. (A and B) Normalized deposited data from 2 GEO EAC cohorts from Kimchi et al29 (GEO series GSE1420) and Wang et al30 (GEO series GSE26886), which support the loss of UBE2D3 expression between BE and EAC, as well as the observed high UBE2D3 expression in both SE and BE. Both studies applied histopathology-driven tissue dissection and U133 Affymetrix expression array platforms, but different analytic methodologies. P values represent unequal variances t tests applied between BE and EAC groups in each cohort. (C) Lack of correlated expression between UBE2D1 and UBE2D3 across the 65 RNAseq samples in the BE progression cohort. (D–G) RNAseq by expectation maximization (RSEM) expression vs capped relative linear copy number plots in TCGA EAC samples (n = 87) from ESCA cohort31 for (D) UBE2D1, (E) UBE2D3, (F) UBE2E1, and (G) UBE2I.