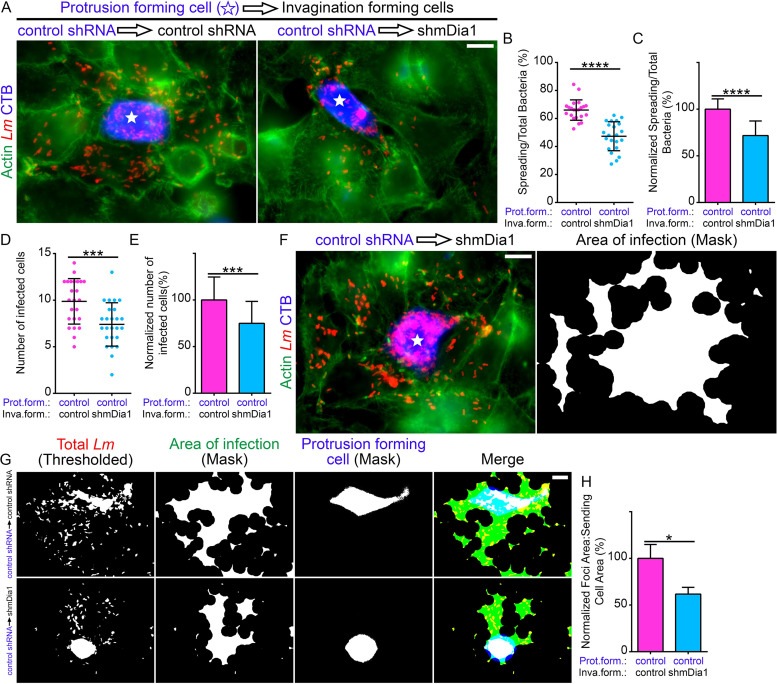
FIG 6.
Optimal L. monocytogenes cell-to-cell spread relies on mDia1 expression in the bacterial membrane invagination-forming cells. (A) Micrographs representative of the mixed-cell spreading assays. Preinfected HeLa cells stably expressing the control (nontargeting) shRNA (control shRNA) and labeled with CellTracker Blue (CTB) (blue) were mixed with uninfected (and unlabeled) control (control shRNA) or mDia1-depleted (shmDia1) cells. Samples were stained with anti-L. monocytogenes antibodies (Lm) (red) and Alexa Fluor 488-phalloidin (“Actin”) (green) to visualize F-actin. Bar, 10 μm. The white stars indicate the locations of the protrusion-forming host cells. (B) Quantification of the proportion of bacteria that spread (into surrounding nonblue invagination-forming [termed “inva.form.” on the graphs] host cells) out of the CellTracker Blue-labeled protrusion-forming [“prot.form.” on the graphs] host cells. Twenty-two infection foci containing a total of 6,961 bacteria from samples containing control shRNA invagination-forming host cells (pink circles) were analyzed. The same number of infection foci (with a total of 7,846 bacteria) containing mDia1-depleted (shmDia1) invagination-forming host cells (blue circles) were also analyzed. The average ratios (expressed as a percent) of spreading bacteria to total bacteria (depicted as a scatterplot [mean ± SD]) are 66.01% (foci with control shRNA invagination-forming cells) and 47.33% (foci with shmDia1 invagination-forming cells). ****, P < 0.0001 (unpaired parametric two-tailed t tests [with Welch’s correction]). (C) The data from panel B but normalized to the control (foci with control shRNA invagination-forming cells). Percent values (relative to control, [mean plus SD]) are 100% (foci with control shRNA invagination-forming cells [pink]) and 71.6979% (foci with shmDia1 invagination-forming cells [blue]). ****, P < 0.0001 (unpaired parametric two-tailed t tests [with Welch’s correction]). (D) Quantification of the number of infected non-CellTracker Blue-labeled cells from the mixed-cell spreading assays. A total of 24 and 25 infection foci containing the control shRNA or shmDia1 invagination-forming cells, respectively, were analyzed. The average number of infected cells (depicted as a scatterplot [mean ± SD]) are 9.875 (foci with control shRNA invagination-forming cells [pink]) and 7.4 (foci with shmDia1 invagination-forming cells [blue]). ***, P < 0.001 (unpaired parametric two-tailed t tests [with Welch’s correction]). (E) The data from panel D but normalized to the control (foci with control shRNA invagination-forming cells). Percent values (relative to control [mean plus SD]) are 100% (foci with control shRNA invagination-forming cells [pink]) and 74.94% (foci with shmDia1 invagination-forming cells [blue]). ***, P < 0.001 (unpaired parametric two-tailed t tests [with Welch’s correction]). (F) An example of an infection focus from a mixed-cell spreading assay where the L. monocytogenes bacteria (red) are highly concentrated in the CellTracker Blue-labeled (blue) infected control (control shRNA) protrusion-forming host cell. Bar, 10 μm. The right image depicts the area of infection based on a mask of the thresholded bacteria. (G and H) Quantification of the area of infection by determining the ratio of the total area of infection (green) to the area of the CellTracker Blue-labeled protrusion-forming host cell (“Protrusion Forming Cell”; blue). A mask of the area of infection was created by thresholding the signal of the antibody-labeled L. monocytogenes (red). (H) The average ratios (normalized and expressed as a percentage) of the infection foci area to sending cell area (depicted as a bar graph [mean plus SD]) are 100% (foci with control shRNA invagination-forming cells [pink]) and 61.7289% (foci with shmDia1 invagination-forming cells [blue]). *, P < 0.05 (P is 0.241 by unpaired Mann-Whitney U test). A total of 10 microscopic field of views and 11 microscopic field of views were analyzed from samples containing control shRNA or shmDia1 invagination-forming cells, respectively.