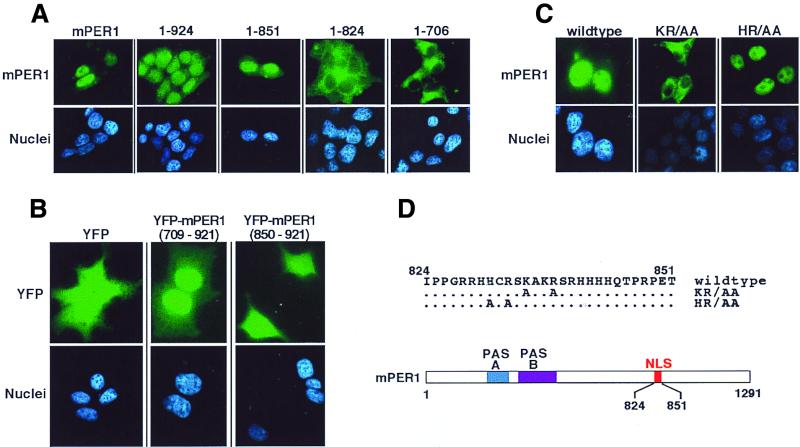
FIG. 5.
Identification of the mPER1 NLS. (A) Progressive truncation of mPER1 reveals a potential NLS. HEK 293 cells were transiently transfected with constructs encoding either full-length Myc-mPER1(mPER1) or one of several carboxyl-terminal truncations of Myc-mPER1 (Fig. 6B). Forty-eight hours posttransfection, the proteins were visualized with Alexa 488 (green)-conjugated anti-Myc MAb 9E10 and the nuclei were visualized with Hoechst staining. Representative micrographs are shown. Quantitation of the nuclear localization of mPER1 and constructs encoding mPER1(1-924) and mPER1(1-851) is shown in Fig. 7. The cytoplasmic localization of mPER1(1-824) and mPER(1-706) was seen in >95% of transfected cells. (B) mPER1 amino acids 709 to 921 are sufficient to direct nuclear localization. HEK 293 cells were transiently transfected with constructs encoding double YFP, either alone or fused to mPER1(709-921) or mPER1(850-921). Double YFP alone and YFP-mPER1(850-921) were diffusely distributed in >95% of transfected cells, while YFP-mPER1(709-921) concentrated in the nuclei of >95% of transfected cells. (C and D) Point mutations identify essential residues in mPER1 NLS. HEK 293 cells were transiently transfected with constructs encoding wild-type or double point mutant K835A/R838A (KR/AA) or H831A/R833A (HR/AA) Myc-mPER1. After transfection, the epitope-tagged mPER1 was visualized as for panel A. A related NLS mutant, K837E/R838D-mPER1, was also cytoplasmic (data not shown). (D) Cartoon demonstrating location of NLS in mPER1 and mutations introduced to create KR/AA and HR/AA Myc-mPER1.