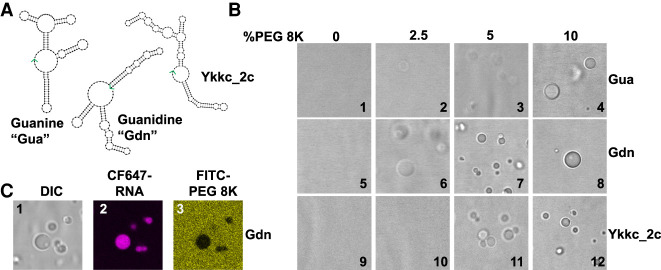
FIGURE 1.
Macromolecular crowding induces condensate formation of bacterial riboswitches. (A) Secondary structure models of guanine (Gua), guanidine (Gdn), and ykkc_2c riboswitches based on NUPACK (Zadeh et al. 2011). See Supplemental Figure S1 for sequences. The 3′ ends of the RNAs are indicated with green arrows. (B) DIC microscope images of riboswitch RNA condensates in the presence of polyethylene glycol 8K (PEG8K). 10 µM RNA was renatured in the presence of H10N15M10 buffer (see Materials and Methods), and varying amounts of polyethylene glycol (8K) for the indicated riboswitches. Renaturation was carried out by heating the mixture at 90°C for 3 min, 65°C for 1 min, and holding at 37°C until mounted on a glass slide with DIC visualization. (C) Condensates contain RNA and are depleted in PEG. Conditions as per image 8, except CF647-labeled guanidine riboswitch (8% labeled) and FITC-labeled PEG-8K (0.5% labeled) were used in all three images. See Materials and Methods for excitation and emission parameters.