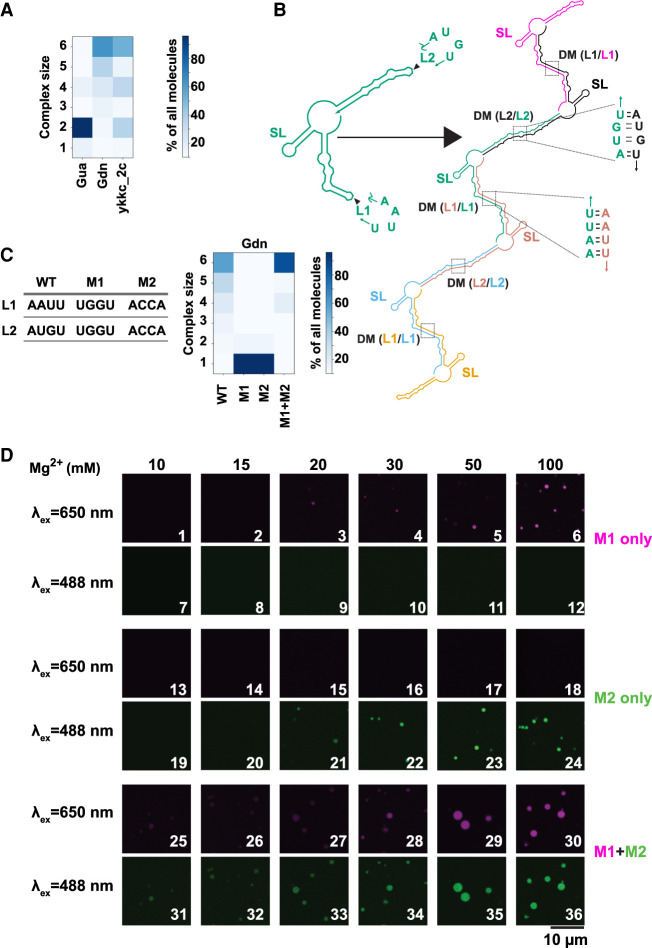
FIGURE 3.
Loop–loop interactions mediate assembly of RNA condensates. (A) Propensity for RNA multimerization as predicted by NUPACK for guanine (Gua), guanidine (Gdn), and ykkc_2c nucleotide binding riboswitch (ykkc_2c). (B) NUPACK predicted architecture for multimerization (n = 6 here) of guanidine riboswitch. Self-complementary loops L1 and L2 form trans-strand dimerization motif (DM). Stem–loop structure (SL) is retained as a cis-strand structure within the multimers. Each copy of the transcript is a separate color and labels are color matched. (C) Propensity for RNA multimerization of the guanidine riboswitch as predicted by NUPACK for WT, M1, M2, and M1 + M2 mutants. M1 was CF647 labeled and M2 was CF488 labeled. (D) Guanidine riboswitch mutant RNAs were renatured with indicated amounts of Mg2+ (in the absence of PEG). Samples contained 2.5 µM total RNA, with ∼8% labeled with indicated dyes. For experiments containing both M1 and M2, half amounts of individual RNAs were premixed while keeping the total RNA concentrations identical to experiments containing single RNA only.