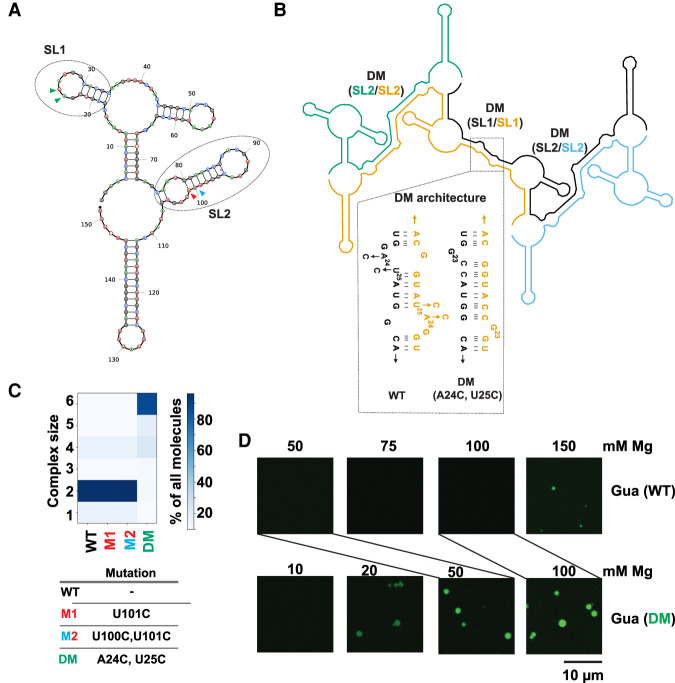
FIGURE 4.
Stabilization of RNA–RNA interactions favors LLPS of the Guanine riboswitch. (A) Predicted secondary structure of the Guanine riboswitch monomer. Regions containing unpaired nucleotides that become part of base-paired structures post-multimerization are indicated as SL1 and SL2 and encompassed with ovals. Colored arrows refer to positions of mutants below. (B) NUPACK model for multimerization of the Guanine riboswitch (n = 4 here). Self-complementary SL1 and SL2 form extended dimer helices (DM) in trans. The SL1 region was stabilized by the A24C:U25C double mutant designed to stabilize DM (SL1/SL1) region (shown in dotted region). Each copy of the transcript is a separate color and labels are color-matched. (C) The SL2 region was stabilized by the M1 and M2 changes (SL2/SL2). See panel A for color-matching arrows. (D). Microscopy images comparing formation of condensates by the WT (10% labeled RNA) and A24C:U25C double mutant guanine (15% labeled RNA) riboswitch. Samples contained 2.5 µM RNA in 10 mM HEPES and 15 mM NaCl (pH 7.0) with indicated concentrations of Mg2+. Lines directly compare the 50 mM Mg2+ concentrations for WT and DM, as well as the 100 mM Mg2+ concentrations.