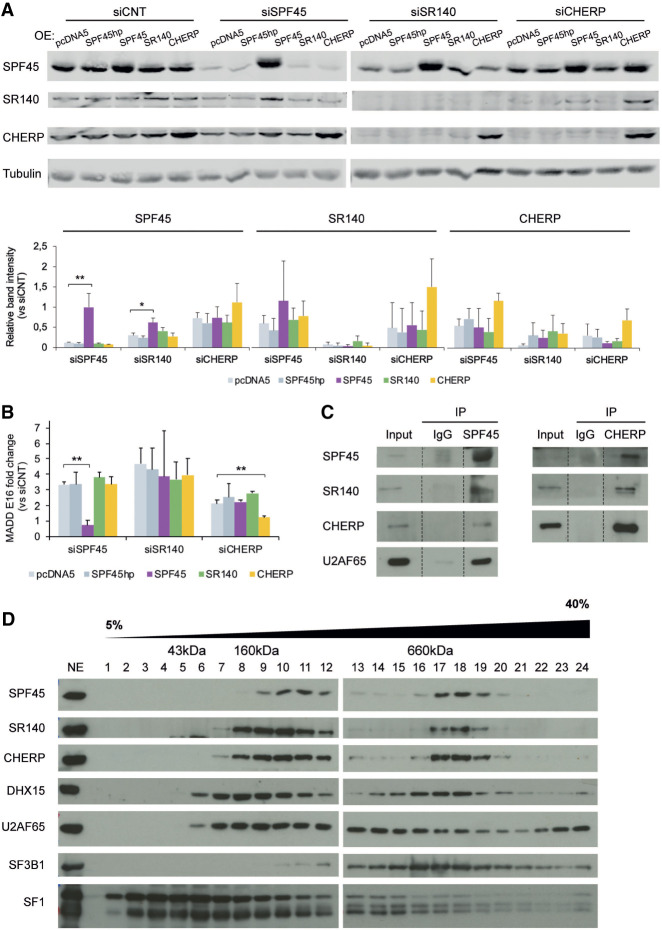
FIGURE 2.
SPF45, SR140, and CHERP form a functional physical complex. (A, upper panel) Western blot analysis of SPF45, SR140, and CHERP (and tubulin as loading control) upon 3′UTR-targeted siRNA knockdowns in Flp-In T-Rex HeLa cells stably overexpressing either an empty vector (pcDNA5), a SPF45 cDNA containing a hairpin to prevent translation (SPF45hp), or cDNAs of SPF45, SR140, or CHERP with 3′UTRs not targeted by the siRNAs utilized. Twenty-four hours after siRNA transfection, protein expression was induced using tetracycline, and total cell extracts were prepared 48 h later. The results shown are representative of three independent experiments. (Lower panel) Quantification of western blot results, represented as mean ± SD of three biological replicates. P-values calculated comparing to pcDNA5 by one-way ANOVA followed by Dunnett's multiple comparison test ([*] P < 0.01; [**] P < 0.001). (B) Quantification of MADD exon 16 AS by RT-PCR followed by capillary electrophoresis, represented as fold change compared to siCNT in the same experimental conditions of A. Data are represented as mean ± SD of three biological replicates, and P-values are calculated comparing to pcDNA5 by Student's t-test ([**] P < 0.01). (C) Immunoprecipitation of endogenous SPF45 or CHERP in total protein extracts from HeLa cells and detection of coimmunoprecipitating proteins by western blot analyses. (D) Fractionation of HeLa nuclear extract (NE) through a 5%–40% glycerol gradient followed by western blot analysis of the indicated proteins. The approximate size of the fractions was estimated by profiling a molecular weight marker kit through a parallel gradient and staining it with Coomassie. The result shown is representative of three independent experiments.