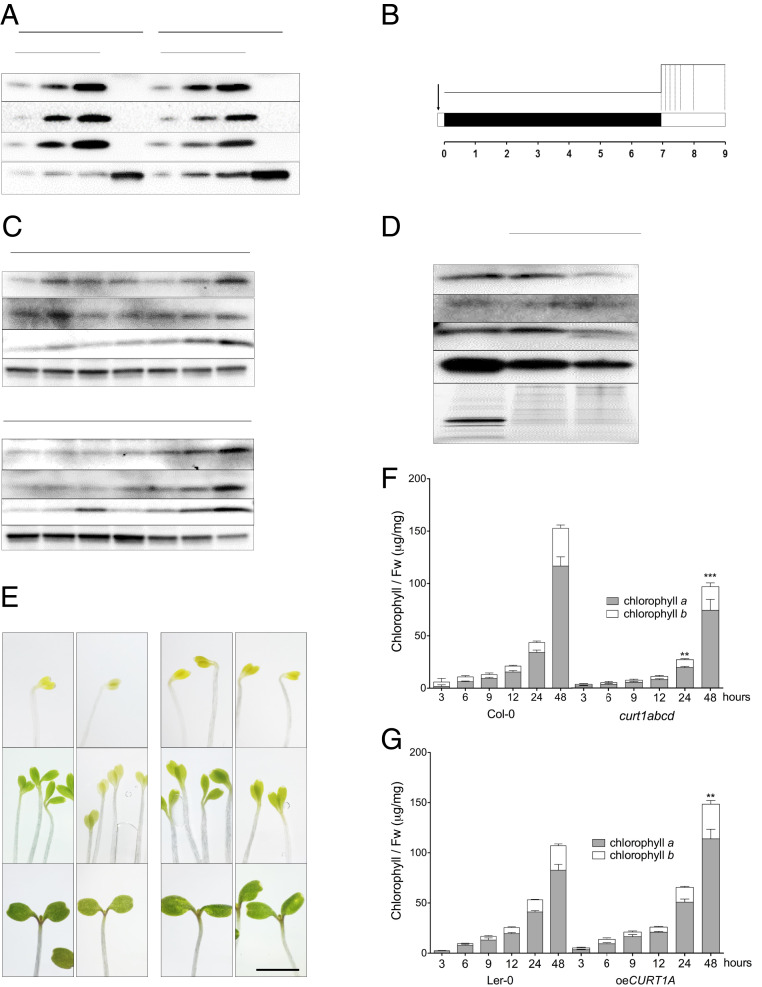
Fig. 1.
CURT1 proteins are present in membrane fractions of cotyledons throughout de-etiolation. (A) Accumulation of CURT1A-C proteins in cotyledons and roots of 7-d-old Col-0 and Ler-0 seedlings grown under a 16/8 h light/dark cycle. ACTIN was used as the loading control. (B) Design of the de-etiolation assay. Seeds were stratified for 2 to 3 d and exposed to light for 1 h prior to dark acclimation for 1 wk. Seedlings were sampled at 0, 3, 6, 9, 12, 24, and 48 h after the onset of continuous light (dotted lines). (C) Immunoblotting analysis of CURT1A-C accumulation in total protein extracts from seedlings sampled at the times shown in B; 10 μg of protein was loaded in each lane. n = 3. (D) Membrane fractions (M. fractions) were prepared from cotyledons after 6 h of illumination. The accumulation of CURT1A-C proteins in mature thylakoids (T) and membrane fractions was analyzed in Col-0 and Ler-0. AtpB, a typical integral membrane protein, served as the loading control. n = 3. (E) Images of emerging cotyledons from Col-0, curt1abcd, Ler-0, and oeCURT1A at 0, 12, and 48 h after the onset of illumination. (Scale bar, 5 mm.) (F and G) Total chlorophyll content analyzed in (F) Col-0, curt1abcd, and (G) Ler-0 and oeCURT1A after 3, 6, 9, 12, 24, and 48 h of de-etiolation. The contributions of chlorophyll a (black columns) and chlorophyll b (white columns) to the total chlorophyll content are depicted. Error bars represent SDs of six biological replicates. Levels at 48 h were compared (by two-way ANOVA with Bonferroni posttest) between curt1abcd and Col-0 and between oeCURT1A and Ler-0. **P < 0.01; ***P < 0.001.