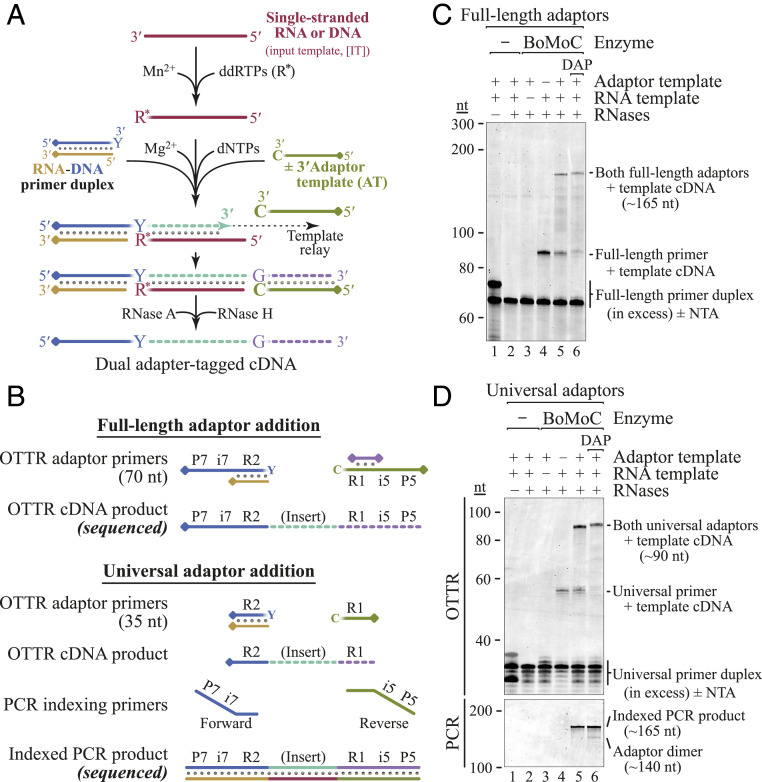
Fig. 3.
OTTR for NGS cDNA library generation. (A) Optimized workflow for single-tube synthesis of cDNA libraries. A pool of RNA and/or DNA input molecules (maroon) is first labeled by BoMoC with 3′ ddRTP. Buffer conditions are then toggled from Mn2+ to Mg2+ and any free ddRTPs are inactivated. Next, dNTPs, oligonucleotides, and BoMoC are added to initiate cDNA synthesis from the RNA-DNA primer duplex across the IT (maroon), ending after copying the AT (green). If desired, products can then be treated with RNase A and RNase H to remove RNA, yielding the desired cDNA. The illustrated blocking groups are detailed in SI Appendix, Table S1. (B) Schematic of primers involved in Illumina Full-length (Top) or Universal (Bottom) adaptor addition and their respective cDNA library products. DNA primers were the complement of P7-i7-R2 or R2, while ATs were P5-i5-R1 or R1. In the Full-length adaptor strategy, only cDNA products elongated by copying the AT can bind to the flow cell. The covalently linked blocking group is indicated by a diamond. (C and D) Proof of principle for OTTR library generation using an RNA oligonucleotide template with Full-length (C) or Universal (D) adaptors. All reactions contained primer duplex. Only reactions containing primer duplex, RNA template, AT, and BoMoC (lanes 5 and 6) generate properly sized cDNA library product. Universal adaptor RT reactions required PCR amplification for P5 and P7 sequence fusion and indexing (D, Bottom). DAP, 2-amino-2′-deoxyadenosine triphosphate.