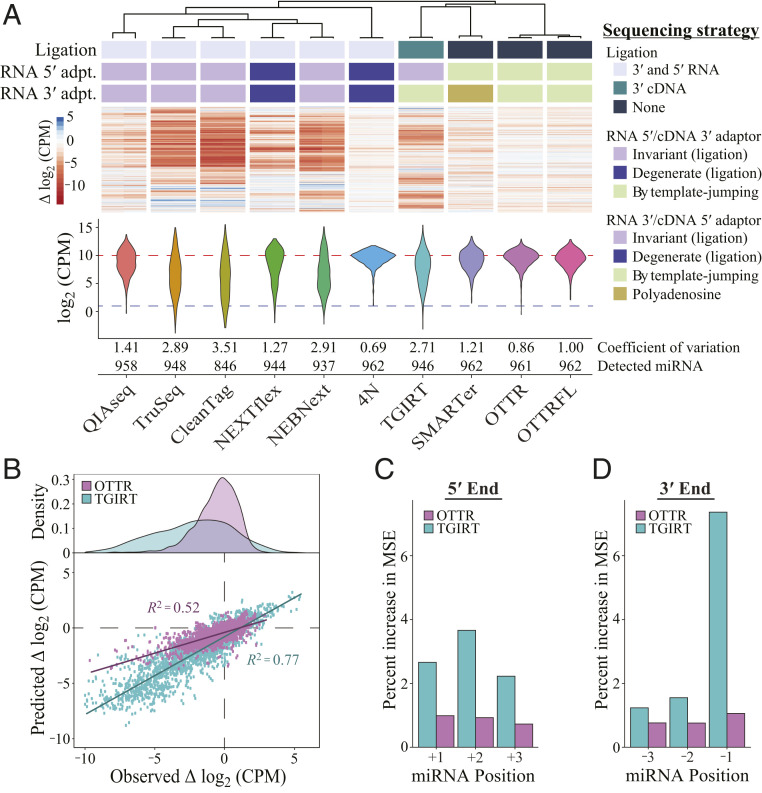
Fig. 4.
OTTR outperforms library generation protocols from commercially available kits. Sequencing reads other than from OTTR were sampled by downloading published datasets (35, 38–40). (A) Unsupervised hierarchical clustering of ∆log2CPM of miRNA read counts from libraries made using different protocols, with side-by-side paired technical replicates for each. ∆log2CPM = log2 (CPMexpected/CPMobserved), where CPMexpected is 962−1 × 1,000,000. The dendrogram indicates the relatedness of miRNA read-count bias. Annotations below the dendrogram indicate protocol distinctions in ligase and polymerase usage. Ligated adaptors were considered “degenerate” or “invariant” based on whether the adapter sequence had mixed-base positions. “Polyadenosine” indicates tailing of the input RNA by polyA polymerase necessary for binding of an oligothymidine RT primer. DESeq2 was used to normalize read counts for each set of replicates before conversion to log2CPM, and the distributions for combined replicates are presented as violin plots. Across the violins, the red dashed line defines the expected mean log2CPM of equimolar representation, and the blue dashed line defines the detection cutoff, which was CPM > 2. (B) Evaluation of random forest models’ predicted ∆log2CPM and observed ∆log2CPM for each miRNA based on the 5′-most (+1, +2, and +3) or 3′-most (−3, −2, −1) bases. (C and D) Percent increase in mean squared error (MSE) or relative importance for each variable of the random forest model trained on OTTR and TGIRT datasets (C: +1, +2, and +3 for the three 5′-most bases where +1 is the exact 5′ end; D: −3, −2, and −1 for the three 3′-most bases where −1 is the exact 3′ end). Variables with a higher-percent MSE are considered more important in the random forest model when predicting the log2CPM.